Identifying FDA-approved drugs with multimodal properties against COVID-19 using a data-driven approach and a lung organoid model of SARS-CoV-2 entry
- PMID: 34503440
- PMCID: PMC8426591
- DOI: 10.1186/s10020-021-00356-6
Identifying FDA-approved drugs with multimodal properties against COVID-19 using a data-driven approach and a lung organoid model of SARS-CoV-2 entry
Abstract
Background: Vaccination programs have been launched worldwide to halt the spread of COVID-19. However, the identification of existing, safe compounds with combined treatment and prophylactic properties would be beneficial to individuals who are waiting to be vaccinated, particularly in less economically developed countries, where vaccine availability may be initially limited.
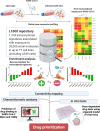
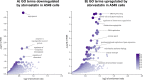
Methods: We used a data-driven approach, combining results from the screening of a large transcriptomic database (L1000) and molecular docking analyses, with in vitro tests using a lung organoid model of SARS-CoV-2 entry, to identify drugs with putative multimodal properties against COVID-19.
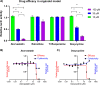
Results: Out of thousands of FDA-approved drugs considered, we observed that atorvastatin was the most promising candidate, as its effects negatively correlated with the transcriptional changes associated with infection. Atorvastatin was further predicted to bind to SARS-CoV-2's main protease and RNA-dependent RNA polymerase, and was shown to inhibit viral entry in our lung organoid model.
Conclusions: Small clinical studies reported that general statin use, and specifically, atorvastatin use, are associated with protective effects against COVID-19. Our study corroborrates these findings and supports the investigation of atorvastatin in larger clinical studies. Ultimately, our framework demonstrates one promising way to fast-track the identification of compounds for COVID-19, which could similarly be applied when tackling future pandemics.
Keywords: Atorvastatin; COVID-19; Chemoinformatics; Connectivity mapping; Drug repositioning; Drug screening; Drug testing; Lung organoids; Molecular docking; Pandemic.
© 2021. The Author(s).
Conflict of interest statement
RES is on the scientific advisory board of Miromatrix Inc. and is a consultant and speaker for Alnylam Inc. The other authors declare no competing interests.
Figures

References
-
- Bowers KJ, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In: Proceedings of the 2006 ACM/IEEE conference on supercomputing. 2006. Pp. 43–43. 10.1109/SC.2006.54
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

