Phenotypic Expression and Outcomes in Individuals With Rare Genetic Variants of Hypertrophic Cardiomyopathy
- PMID: 34503678
- PMCID: PMC8434420
- DOI: 10.1016/j.jacc.2021.07.017
Phenotypic Expression and Outcomes in Individuals With Rare Genetic Variants of Hypertrophic Cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is caused by rare variants in sarcomere-encoding genes, but little is known about the clinical significance of these variants in the general population.
Objectives: The goal of this study was to compare lifetime outcomes and cardiovascular phenotypes according to the presence of rare variants in sarcomere-encoding genes among middle-aged adults.
Methods: This study analyzed whole exome sequencing and cardiac magnetic resonance imaging in UK Biobank participants stratified according to sarcomere-encoding variant status.
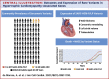
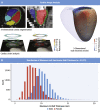
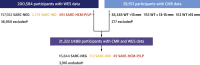
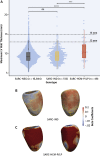
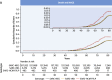
Results: The prevalence of rare variants (allele frequency <0.00004) in HCM-associated sarcomere-encoding genes in 200,584 participants was 2.9% (n = 5,712; 1 in 35), and the prevalence of variants pathogenic or likely pathogenic for HCM (SARC-HCM-P/LP) was 0.25% (n = 493; 1 in 407). SARC-HCM-P/LP variants were associated with an increased risk of death or major adverse cardiac events compared with controls (hazard ratio: 1.69; 95% confidence interval [CI]: 1.38-2.07; P < 0.001), mainly due to heart failure endpoints (hazard ratio: 4.23; 95% CI: 3.07-5.83; P < 0.001). In 21,322 participants with both cardiac magnetic resonance imaging and whole exome sequencing, SARC-HCM-P/LP variants were associated with an asymmetric increase in left ventricular maximum wall thickness (10.9 ± 2.7 mm vs 9.4 ± 1.6 mm; P < 0.001), but hypertrophy (≥13 mm) was only present in 18.4% (n = 9 of 49; 95% CI: 9%-32%). SARC-HCM-P/LP variants were still associated with heart failure after adjustment for wall thickness (hazard ratio: 6.74; 95% CI: 2.43-18.7; P < 0.001).
Conclusions: In this population of middle-aged adults, SARC-HCM-P/LP variants have low aggregate penetrance for overt HCM but are associated with an increased risk of adverse cardiovascular outcomes and an attenuated cardiomyopathic phenotype. Although absolute event rates are low, identification of these variants may enhance risk stratification beyond familial disease.
Keywords: cardiovascular magnetic resonance; deep learning; genetics; hypertrophic cardiomyopathy; penetrance.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures This study was supported by the Medical Research Council, UK (MC-A658-5QEB0); the National Institute for Health Research Imperial College Biomedical Research Centre; the National Institute for Health Research Royal Brompton Cardiovascular Biomedical Research Unit; the British Heart Foundation (NH/17/1/32725, RG/19/6/34387, RE/18/4/34215); Fondation Leducq (16 CVD 03); Wellcome Trust (107469/Z/15/Z, 200990/A/16/Z); the National Heart and Lung Institute Foundation; the Royston Centre for Cardiomyopathy Research; Rosetrees and CORDA (Dr Prasad); Academy of Medical Sciences (SGL015/1006; Dr de Marvao); Mason Medical Research Trust grant (Dr de Marvao); SmartHeart EPSRC Programme Grant (EP/P001009/1; Dr Bai and Dr Rueckert); and a Rosetrees and Stoneygate Imperial College Research Fellowship (Dr Whiffin). Dr Ware has consulted for MyoKardia, Inc. and Foresite Labs. Dr Cook holds shares in Enleofen Bio Pte. Ltd. Dr O’Regan has consulted for Bayer AG. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures


Comment in
-
Hypertrophic Cardiomyopathy in the General Population: Leveraging the UK Biobank Database and Machine Learning Phenotyping.J Am Coll Cardiol. 2021 Sep 14;78(11):1111-1113. doi: 10.1016/j.jacc.2021.07.036. J Am Coll Cardiol. 2021. PMID: 34503679 No abstract available.
References
-
- Elliott P.M., Anastasakis A., Borger M.A. 2014 ESC guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;35:2733–2779. - PubMed
-
- Ommen S.R., Mital S., Burke M.A. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2020;76:e159–e240. - PubMed
-
- Maron B.J., Maron M.S., Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. - PubMed
-
- Miller D.T., Lee K., Gordon A.S. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23:1391–1398. - PubMed
Publication types
MeSH terms
Grants and funding
- MC_U120085815/MRC_/Medical Research Council/United Kingdom
- FS/15/81/31817/BHF_/British Heart Foundation/United Kingdom
- MC_UP_1102/19/MRC_/Medical Research Council/United Kingdom
- MC-A658-5QEB0/MRC_/Medical Research Council/United Kingdom
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- RG/19/6/34387/BHF_/British Heart Foundation/United Kingdom
- RE/18/4/34215/BHF_/British Heart Foundation/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 200990/A/16/Z/WT_/Wellcome Trust/United Kingdom
- NH/17/1/32725/BHF_/British Heart Foundation/United Kingdom
- DH_/Department of Health/United Kingdom
- FS/ICRF/21/26019/BHF_/British Heart Foundation/United Kingdom
- MC_UP_1605/13/MRC_/Medical Research Council/United Kingdom
- MC_UP_1102/20/MRC_/Medical Research Council/United Kingdom
- 107469/Z/15/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

