Comparative genomics provides insights into the aquatic adaptations of mammals
- PMID: 34503999
- PMCID: PMC8449357
- DOI: 10.1073/pnas.2106080118
Comparative genomics provides insights into the aquatic adaptations of mammals
Abstract
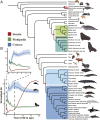
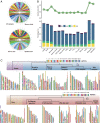
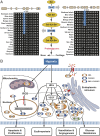
The ancestors of marine mammals once roamed the land and independently committed to an aquatic lifestyle. These macroevolutionary transitions have intrigued scientists for centuries. Here, we generated high-quality genome assemblies of 17 marine mammals (11 cetaceans and six pinnipeds), including eight assemblies at the chromosome level. Incorporating previously published data, we reconstructed the marine mammal phylogeny and population histories and identified numerous idiosyncratic and convergent genomic variations that possibly contributed to the transition from land to water in marine mammal lineages. Genes associated with the formation of blubber (NFIA), vascular development (SEMA3E), and heat production by brown adipose tissue (UCP1) had unique changes that may contribute to marine mammal thermoregulation. We also observed many lineage-specific changes in the marine mammals, including genes associated with deep diving and navigation. Our study advances understanding of the timing, pattern, and molecular changes associated with the evolution of mammalian lineages adapting to aquatic life.
Keywords: aquatic adaptation; comparative genomics; marine mammals.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

Comment in
-
Evolution of pinniped UCP1 is not linked to aquatic life but to neonatal thermogenesis and body size.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2118431119. doi: 10.1073/pnas.2118431119. Proc Natl Acad Sci U S A. 2022. PMID: 35101988 Free PMC article. No abstract available.
-
Reply to Gaudry et al.: Cross-validation is necessary for the identification of pseudogenes.Proc Natl Acad Sci U S A. 2022 Feb 8;119(6):e2120427119. doi: 10.1073/pnas.2120427119. Proc Natl Acad Sci U S A. 2022. PMID: 35101989 Free PMC article. No abstract available.
References
-
- Lü P., et al. ., Genome encode analyses reveal the basis of convergent evolution of fleshy fruit ripening. Nat. Plants 4, 784–791 (2018). - PubMed
-
- Jefferson T. A., Webber M. A., Pitman R. L., Marine Mammals of the World: A Comprehensive Guide to Their Identification (Elsevier, 2015).
-
- Cahill J. A., et al. ., Genomic evidence of widespread admixture from polar bears into brown bears during the last ice age. Mol. Biol. Evol. 35, 1120–1129 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

