A transcriptome-wide association study identifies susceptibility genes for Parkinson's disease
- PMID: 34504106
- PMCID: PMC8429416
- DOI: 10.1038/s41531-021-00221-7
A transcriptome-wide association study identifies susceptibility genes for Parkinson's disease
Abstract
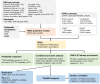
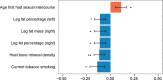
Genome-wide association study (GWAS) has seen great strides in revealing initial insights into the genetic architecture of Parkinson's disease (PD). Since GWAS signals often reside in non-coding regions, relatively few of the associations have implicated specific biological mechanisms. Here, we aimed to integrate the GWAS results with large-scale expression quantitative trait loci (eQTL) in 13 brain tissues to identify candidate causal genes for PD. We conducted a transcriptome-wide association study (TWAS) for PD using the summary statistics of over 480,000 individuals from the most recent PD GWAS. We identified 18 genes significantly associated with PD after Bonferroni corrections. The most significant gene, LRRC37A2, was associated with PD in all 13 brain tissues, such as in the hypothalamus (P = 6.12 × 10-22) and nucleus accumbens basal ganglia (P = 5.62 × 10-21). We also identified eight conditionally independent genes, including four new genes at known PD loci: CD38, LRRC37A2, RNF40, and ZSWIM7. Through conditional analyses, we demonstrated that several of the GWAS significant signals on PD could be driven by genetically regulated gene expression. The most significant TWAS gene LRRC37A2 accounts for 0.855 of the GWAS signal at its loci, and ZSWIM7 accounts for all the GWAS signals at its loci. We further identified several phenotypes previously associated with PD by querying the single nucleotide polymorphisms (SNPs) in the final model of the identified genes in phenome databases. In conclusion, we prioritized genes that are likely to affect PD by using a TWAS approach and identified phenotypes associated with PD.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

