Establishing community reference samples, data and call sets for benchmarking cancer mutation detection using whole-genome sequencing
- PMID: 34504347
- PMCID: PMC8532138
- DOI: 10.1038/s41587-021-00993-6
Establishing community reference samples, data and call sets for benchmarking cancer mutation detection using whole-genome sequencing
Abstract
The lack of samples for generating standardized DNA datasets for setting up a sequencing pipeline or benchmarking the performance of different algorithms limits the implementation and uptake of cancer genomics. Here, we describe reference call sets obtained from paired tumor-normal genomic DNA (gDNA) samples derived from a breast cancer cell line-which is highly heterogeneous, with an aneuploid genome, and enriched in somatic alterations-and a matched lymphoblastoid cell line. We partially validated both somatic mutations and germline variants in these call sets via whole-exome sequencing (WES) with different sequencing platforms and targeted sequencing with >2,000-fold coverage, spanning 82% of genomic regions with high confidence. Although the gDNA reference samples are not representative of primary cancer cells from a clinical sample, when setting up a sequencing pipeline, they not only minimize potential biases from technologies, assays and informatics but also provide a unique resource for benchmarking 'tumor-only' or 'matched tumor-normal' analyses.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
L.T.F., S.M.E.S., M. Mohiyuddin, Y.G., L.Y. and H.L. are employees of Roche Sequencing Solutions Inc. L.K., K.L. and M. Mars are employees of ATCC, which provides cell lines and derivative materials. E.J., G.P.S. and O.D.A. are employees of Illumina Inc. V.P. and M.S. are employees of Novartis Institutes for Biomedical Research. T.H., E.P. and R. Kalamegham are employees of Genentech (a member of the Roche group). Z.L. is an employee of Sentieon Inc. R. Kusko is an employee of Immuneering Corp. C.C., S.M. and J.S. are employees of 10x Genomics. All other authors declare no competing interests.
Figures


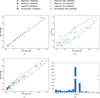


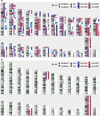








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

