Metagenomic Next-Generation Sequencing of Bloodstream Microbial Cell-Free Nucleic Acid in Children With Suspected Sepsis in Pediatric Intensive Care Unit
- PMID: 34504805
- PMCID: PMC8421769
- DOI: 10.3389/fcimb.2021.665226
Metagenomic Next-Generation Sequencing of Bloodstream Microbial Cell-Free Nucleic Acid in Children With Suspected Sepsis in Pediatric Intensive Care Unit
Abstract
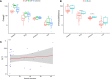
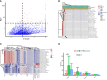
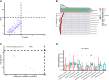
Bloodstream infection is a life-threatening complication in critically ill patients. Multi-drug resistant bacteria or fungi may increase the risk of invasive infections in hospitalized children and are difficult to treat in intensive care units. The purpose of this study was to use metagenomic next-generation sequencing (mNGS) to understand the bloodstream microbiomes of children with suspected sepsis in a pediatric intensive care unit (PICU). mNGS were performed on microbial cell-free nucleic acid from 34 children admitted to PICU, and potentially pathogenic microbes were identified. The associations of serological inflammation indicators, lymphocyte subpopulations, and other clinical phenotypes were also examined. mNGS of blood samples from children in PICU revealed potential eukaryotic microbial pathogens. The abundance of Pneumocystis jirovecii was positively correlated with a decrease in total white blood cell count and immunodeficiency. Hospital-acquired pneumonia patients showed a significant increase in blood bacterial species richness compared with community-acquired pneumonia children. The abundance of bloodstream bacteria was positively correlated with serum procalcitonin level. Microbial genome sequences from potential pathogens were detected in the bloodstream of children with suspected sepsis in PICU, suggesting the presence of bloodstream infections in these children.
Keywords: Pneumocystis jirovecii; bloodstream infection; metagenomic next-generation sequencing; pediatric intensive care unit; sepsis.
Copyright © 2021 Yan, Liu, Chen, Chen, Cheng, Tao, Cai, Zhou, Wang, Wang and Lu.
Conflict of interest statement
Author YZ was employed by company BGI PathoGenesis Pharmaceutical Technology Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Comparison of metagenomic next-generation sequencing and blood culture for diagnosis of bloodstream infections.Front Cell Infect Microbiol. 2024 Jan 24;14:1338861. doi: 10.3389/fcimb.2024.1338861. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38328669 Free PMC article.
-
Metagenomic next-generation sequencing of bronchoalveolar lavage fluid from children with severe pneumonia in pediatric intensive care unit.Front Cell Infect Microbiol. 2023 Mar 16;13:1082925. doi: 10.3389/fcimb.2023.1082925. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37009495 Free PMC article.
-
Comprehensive evaluation of plasma microbial cell-free DNA sequencing for predicting bloodstream and local infections in clinical practice: a multicenter retrospective study.Front Cell Infect Microbiol. 2024 Jan 4;13:1256099. doi: 10.3389/fcimb.2023.1256099. eCollection 2023. Front Cell Infect Microbiol. 2024. PMID: 38362158 Free PMC article.
-
Metagenomics to Assist in the Diagnosis of Bloodstream Infection.J Appl Lab Med. 2019 Jan;3(4):643-653. doi: 10.1373/jalm.2018.026120. Epub 2018 Nov 30. J Appl Lab Med. 2019. PMID: 31639732 Review.
-
Navigating Clinical Utilization of Direct-from-Specimen Metagenomic Pathogen Detection: Clinical Applications, Limitations, and Testing Recommendations.Clin Chem. 2020 Nov 1;66(11):1381-1395. doi: 10.1093/clinchem/hvaa183. Clin Chem. 2020. PMID: 33141913 Review.
Cited by
-
Clinical Evaluation of Metagenomic Next-Generation Sequencing for the detection of pathogens in BALF in severe community acquired pneumonia.Ital J Pediatr. 2023 Feb 18;49(1):25. doi: 10.1186/s13052-023-01431-w. Ital J Pediatr. 2023. PMID: 36805803 Free PMC article.
-
Direct prediction of antimicrobial resistance in Pseudomonas aeruginosa by metagenomic next-generation sequencing.Front Microbiol. 2024 Jun 6;15:1413434. doi: 10.3389/fmicb.2024.1413434. eCollection 2024. Front Microbiol. 2024. PMID: 38903781 Free PMC article.
-
Diagnostic performance and clinical impact of blood metagenomic next-generation sequencing in ICU patients suspected monomicrobial and polymicrobial bloodstream infections.Front Cell Infect Microbiol. 2023 Jun 26;13:1192931. doi: 10.3389/fcimb.2023.1192931. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37434786 Free PMC article.
-
Metagenomics by next-generation sequencing (mNGS) in the etiological characterization of neonatal and pediatric sepsis: A systematic review.Front Pediatr. 2023 Mar 30;11:1011723. doi: 10.3389/fped.2023.1011723. eCollection 2023. Front Pediatr. 2023. PMID: 37063664 Free PMC article. Review.
-
Comparison of metagenomic next-generation sequencing and blood culture for diagnosis of bloodstream infections.Front Cell Infect Microbiol. 2024 Jan 24;14:1338861. doi: 10.3389/fcimb.2024.1338861. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38328669 Free PMC article.
References
-
- Armstrong A. E., Rossoff J., Hollemon D., Hong D. K., Muller W. J., Chaudhury S. (2019). Cell-Free DNA Next-Generation Sequencing Successfully Detects Infectious Pathogens in Pediatric Oncology and Hematopoietic Stem Cell Transplant Patients at Risk for Invasive Fungal Disease. Pediatr. Blood Cancer 66 (7), e27734. 10.1002/pbc.27734 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

