Complement activation promoted by the lectin pathway mediates C3aR-dependent sarcoma progression and immunosuppression
- PMID: 34505065
- PMCID: PMC8425276
- DOI: 10.1038/s43018-021-00173-0
Complement activation promoted by the lectin pathway mediates C3aR-dependent sarcoma progression and immunosuppression
Abstract
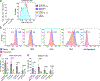
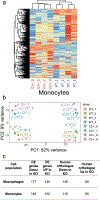
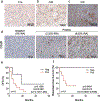
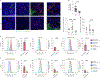
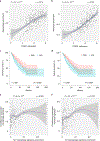
Complement has emerged as a component of tumor promoting inflammation. We conducted a systematic assessment of the role of complement activation and effector pathways in sarcomas. C3-/-, MBL1/2-/- and C4-/- mice showed reduced susceptibility to 3-methylcholanthrene sarcomagenesis and transplanted sarcomas, whereas C1q and factor B deficiency had marginal effects. Complement 3a receptor (C3aR), but not C5aR1 and C5aR2, deficiency mirrored the phenotype of C3-/- mice. C3 and C3aR deficiency were associated with reduced accumulation and functional skewing of tumor-associated macrophages, increased T cell activation and response to anti-PD-1 therapy. Transcriptional profiling of sarcoma infiltrating macrophages and monocytes revealed the enrichment of MHC II-dependent antigen presentation pathway in C3-deficient cells. In patients, C3aR expression correlated with a macrophage population signature and C3 deficiency-associated signatures predicted better clinical outcome. These results suggest that the lectin pathway and C3a/C3aR axis are key components of complement and macrophage-mediated sarcoma promotion and immunosuppression.
Keywords: C3; C3a receptor (C3aR); Complement; cancer; cancer-related inflammation; immunotherapy; lectin pathway; macrophages; mannose binding lectin (MBL); sarcoma.
Conflict of interest statement
Declaration of interests The authors except J.D.L. declare no competing financial interests. J.D.L. is the founder of Amyndas Pharmaceuticals, which is developing complement inhibitors for therapeutic purposes, is the inventor of patents or patent applications that describe the use of complement inhibitors for therapeutic purposes, some of which are developed by Amyndas Pharmaceuticals. J.D.L. is also the inventor of the compstatin technology licensed to Apellis Pharmaceuticals [i.e., 4(1MeW)7W/POT-4/APL-1 and PEGylated derivatives such as APL-2/pegcetacoplan].
Figures




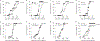




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

