Prioritization and functional analysis of GWAS risk loci for Barrett's esophagus and esophageal adenocarcinoma
- PMID: 34505128
- PMCID: PMC8825357
- DOI: 10.1093/hmg/ddab259
Prioritization and functional analysis of GWAS risk loci for Barrett's esophagus and esophageal adenocarcinoma
Abstract
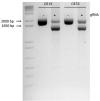
Genome-wide association studies (GWAS) have identified ~20 genetic susceptibility loci for esophageal adenocarcinoma (EAC), and its precursor, Barrett's esophagus (BE). Despite such advances, functional/causal variants and gene targets at these loci remain undefined, hindering clinical translation. A key challenge is that most causal variants map to non-coding regulatory regions such as enhancers, and typically, numerous potential candidate variants at GWAS loci require testing. We developed a systematic informatics pipeline for prioritizing candidate functional variants via integrative functional potential scores (FPS) consolidated from multi-omics annotations, and used this pipeline to identify two high-scoring variants for experimental interrogation: chr9q22.32/rs11789015 and chr19p13.11/rs10423674. Minimal candidate enhancer regions spanning these variants were evaluated using luciferase reporter assays in two EAC cell lines. One of the two variants tested (rs10423674) exhibited allele-specific enhancer activity. CRISPR-mediated deletion of the putative enhancer region in EAC cell lines correlated with reduced expression of two genes-CREB-regulated transcription coactivator 1 (CRTC1) and Cartilage oligomeric matrix protein (COMP); expression of five other genes remained unchanged (CRLF1, KLHL26, TMEM59L, UBA52, RFXANK). Expression quantitative trait locus mapping indicated that rs10423674 genotype correlated with CRTC1 and COMP expression in normal esophagus. This study represents the first experimental effort to bridge GWAS associations to biology in BE/EAC and supports the utility of FPS to guide variant prioritization. Our findings reveal a functional variant and candidate risk enhancer at chr19p13.11 and implicate CRTC1 and COMP as putative gene targets, suggesting that altered expression of these genes may underlie the BE/EAC risk association.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures






References
-
- Coleman, H.G., Xie, S.-H. and Lagergren, J. (2017) The epidemiology of esophageal adenocarcinoma. Gastroenterology, 154, 390–405. - PubMed
-
- Naef, A.P., Savary, M. and Ozzello, L. (1975) Columnar-lined lower esophagus: an acquired lesion with malignant predisposition. Report on 140 cases of Barrett’s esophagus with 12 adenocarcinomas. J. Thorac. Cardiovasc. Surg., 70, 826–835. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

