A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria
- PMID: 34505573
- PMCID: PMC8460266
- DOI: 10.7554/eLife.72409
A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria
Abstract
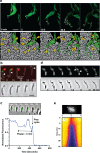
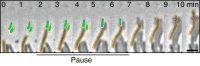
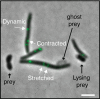
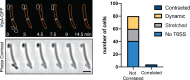
Myxococcus xanthus, a soil bacterium, predates collectively using motility to invade prey colonies. Prey lysis is mostly thought to rely on secreted factors, cocktails of antibiotics and enzymes, and direct contact with Myxococcus cells. In this study, we show that on surfaces the coupling of A-motility and contact-dependent killing is the central predatory mechanism driving effective prey colony invasion and consumption. At the molecular level, contact-dependent killing involves a newly discovered type IV filament-like machinery (Kil) that both promotes motility arrest and prey cell plasmolysis. In this process, Kil proteins assemble at the predator-prey contact site, suggesting that they allow tight contact with prey cells for their intoxication. Kil-like systems form a new class of Tad-like machineries in predatory bacteria, suggesting a conserved function in predator-prey interactions. This study further reveals a novel cell-cell interaction function for bacterial pili-like assemblages.
Keywords: Myxococcus xanthus; infectious disease; microbiology; motility; predation; tad pilus.
© 2021, Seef et al.
Conflict of interest statement
SS, JH, Pd, LM, GB, DR, RJ, RM, EC, BH No competing interests declared, TM Reviewing editor, eLife
Figures







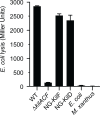
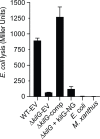
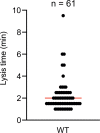

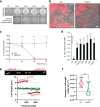
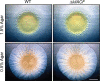
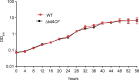

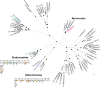

Comment in
-
Contact-dependent killing by Myxococcus.Nat Rev Microbiol. 2021 Dec;19(12):744. doi: 10.1038/s41579-021-00644-2. Nat Rev Microbiol. 2021. PMID: 34588656 No abstract available.
References
-
- Avidan O, Petrenko M, Becker R, Beck S, Linscheid M, Pietrokovski S, Jurkevitch E. Identification and characterization of Differentially-Regulated type IVb pilin genes necessary for predation in obligate bacterial predators. Scientific Reports. 2017;7:1013. doi: 10.1038/s41598-017-00951-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

