Identifying indicator species in ecological habitats using Deep Optimal Feature Learning
- PMID: 34506523
- PMCID: PMC8432828
- DOI: 10.1371/journal.pone.0256782
Identifying indicator species in ecological habitats using Deep Optimal Feature Learning
Abstract
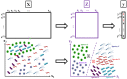
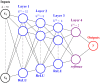
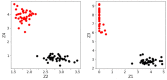
Much of the current research on supervised modelling is focused on maximizing outcome prediction accuracy. However, in engineering disciplines, an arguably more important goal is that of feature extraction, the identification of relevant features associated with the various outcomes. For instance, in microbial communities, the identification of keystone species can often lead to improved prediction of future behavioral shifts. This paper proposes a novel feature extractor based on Deep Learning, which is largely agnostic to underlying assumptions regarding the training data. Starting from a collection of microbial species abundance counts, the Deep Learning model first trains itself to classify the selected distinct habitats. It then identifies indicator species associated with the habitats. The results are then compared and contrasted with those obtained by traditional statistical techniques. The indicator species are similar when compared at top taxonomic levels such as Domain and Phylum, despite visible differences in lower levels such as Class and Order. More importantly, when our estimated indicators are used to predict final habitat labels using simpler models (such as Support Vector Machines and traditional Artificial Neural Networks), the prediction accuracy is improved. Overall, this study serves as a preliminary step that bridges modern, black-box Machine Learning models with traditional, domain expertise-rich techniques.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures














Similar articles
-
Transferability of artificial neural networks for clinical document classification across hospitals: A case study on abnormality detection from radiology reports.J Biomed Inform. 2018 Sep;85:68-79. doi: 10.1016/j.jbi.2018.07.017. Epub 2018 Jul 17. J Biomed Inform. 2018. PMID: 30026067
-
Integrated analysis of machine learning and deep learning in chili pest and disease identification.J Sci Food Agric. 2021 Jul;101(9):3582-3594. doi: 10.1002/jsfa.10987. Epub 2020 Dec 24. J Sci Food Agric. 2021. PMID: 33275806
-
Efficient mapping of crash risk at intersections with connected vehicle data and deep learning models.Accid Anal Prev. 2020 Sep;144:105665. doi: 10.1016/j.aap.2020.105665. Epub 2020 Jul 16. Accid Anal Prev. 2020. PMID: 32683130
-
Machine learning applications to clinical decision support in neurosurgery: an artificial intelligence augmented systematic review.Neurosurg Rev. 2020 Oct;43(5):1235-1253. doi: 10.1007/s10143-019-01163-8. Epub 2019 Aug 17. Neurosurg Rev. 2020. PMID: 31422572
-
Protein secondary structure prediction using neural networks and deep learning: A review.Comput Biol Chem. 2019 Aug;81:1-8. doi: 10.1016/j.compbiolchem.2019.107093. Epub 2019 Aug 12. Comput Biol Chem. 2019. PMID: 31442779 Review.
Cited by
-
Enhancing infectious disease prediction model selection with multi-objective optimization: an empirical study.PeerJ Comput Sci. 2024 Jul 29;10:e2217. doi: 10.7717/peerj-cs.2217. eCollection 2024. PeerJ Comput Sci. 2024. PMID: 39145229 Free PMC article.
References
-
- Rajput DS, Basha SM, Xin Q, Gadekallu TR, Kaluri R, Lakshmanna K, et al.. Providing diagnosis on diabetes using cloud computing environment to the people living in rural areas of India. Journal of Ambient Intelligence and Humanized Computing. 2021; p. 1–12.
-
- Podani J, Csányi B. Detecting indicator species: Some extensions of the IndVal measure. Ecological Indicators. 2010;10(6):1119–1124. doi: 10.1016/j.ecolind.2010.03.010 - DOI
-
- Penczak T. Fish assemblage compositions after implementation of the IndVal method on the Narew River system. Ecological modelling. 2009;220(3):419–423. doi: 10.1016/j.ecolmodel.2008.11.005 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

