Robust neuromorphic coupled oscillators for adaptive pacemakers
- PMID: 34508121
- PMCID: PMC8433448
- DOI: 10.1038/s41598-021-97314-3
Robust neuromorphic coupled oscillators for adaptive pacemakers
Abstract
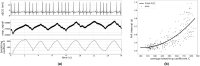
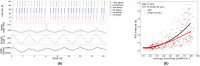
Neural coupled oscillators are a useful building block in numerous models and applications. They were analyzed extensively in theoretical studies and more recently in biologically realistic simulations of spiking neural networks. The advent of mixed-signal analog/digital neuromorphic electronic circuits provides new means for implementing neural coupled oscillators on compact, low-power, spiking neural network hardware platforms. However, their implementation on this noisy, low-precision and inhomogeneous computing substrate raises new challenges with regards to stability and controllability. In this work, we present a robust, spiking neural network model of neural coupled oscillators and validate it with an implementation on a mixed-signal neuromorphic processor. We demonstrate its robustness showing how to reliably control and modulate the oscillator's frequency and phase shift, despite the variability of the silicon synapse and neuron properties. We show how this ultra-low power neural processing system can be used to build an adaptive cardiac pacemaker modulating the heart rate with respect to the respiration phases and compare it with surface ECG and respiratory signal recordings from dogs at rest. The implementation of our model in neuromorphic electronic hardware shows its robustness on a highly variable substrate and extends the toolbox for applications requiring rhythmic outputs such as pacemakers.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Ermentrout B. An introduction to neural oscillators. In: Ventriglia F, editor. Neural Modeling and Neural Networks. Pergamon Press; 1993.
-
- FitzHugh R. Mathematical models of threshold phenomena in the nerve membrane. Bull. Math. Biophys. 1955;17:257–278. doi: 10.1007/BF02477753. - DOI
-
- Terman D, Wang D. Global competition and local cooperation in a network of neural oscillators. Phys. D: Nonlinear Phenom. 1995;81:148–176. doi: 10.1016/0167-2789(94)00205-5. - DOI
Publication types
LinkOut - more resources
Full Text Sources

