Genetic modeling of GNAO1 disorder delineates mechanisms of Gαo dysfunction
- PMID: 34508586
- PMCID: PMC8863422
- DOI: 10.1093/hmg/ddab235
Genetic modeling of GNAO1 disorder delineates mechanisms of Gαo dysfunction
Abstract
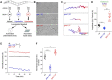
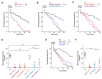
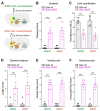
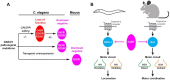
GNAO1 encephalopathy is a neurodevelopmental disorder with a spectrum of symptoms that include dystonic movements, seizures and developmental delay. While numerous GNAO1 mutations are associated with this disorder, the functional consequences of pathological variants are not completely understood. Here, we deployed the invertebrate C. elegans as a whole-animal behavioral model to study the functional effects of GNAO1 disorder-associated mutations. We tested several pathological GNAO1 mutations for effects on locomotor behaviors using a combination of CRISPR/Cas9 gene editing and transgenic overexpression in vivo. We report that all three mutations tested (G42R, G203R and R209C) result in strong loss of function defects when evaluated as homozygous CRISPR alleles. In addition, mutations produced dominant negative effects assessed using both heterozygous CRISPR alleles and transgenic overexpression. Experiments in mice confirmed dominant negative effects of GNAO1 G42R, which impaired numerous motor behaviors. Thus, GNAO1 pathological mutations result in conserved functional outcomes across animal models. Our study further establishes the molecular genetic basis of GNAO1 encephalopathy, and develops a CRISPR-based pipeline for functionally evaluating mutations associated with neurodevelopmental disorders.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

