Emergence of an early SARS-CoV-2 epidemic in the United States
- PMID: 34508652
- PMCID: PMC8313480
- DOI: 10.1016/j.cell.2021.07.030
Emergence of an early SARS-CoV-2 epidemic in the United States
Abstract
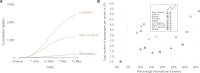
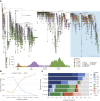
The emergence of the COVID-19 epidemic in the United States (U.S.) went largely undetected due to inadequate testing. New Orleans experienced one of the earliest and fastest accelerating outbreaks, coinciding with Mardi Gras. To gain insight into the emergence of SARS-CoV-2 in the U.S. and how large-scale events accelerate transmission, we sequenced SARS-CoV-2 genomes during the first wave of the COVID-19 epidemic in Louisiana. We show that SARS-CoV-2 in Louisiana had limited diversity compared to other U.S. states and that one introduction of SARS-CoV-2 led to almost all of the early transmission in Louisiana. By analyzing mobility and genomic data, we show that SARS-CoV-2 was already present in New Orleans before Mardi Gras, and the festival dramatically accelerated transmission. Our study provides an understanding of how superspreading during large-scale events played a key role during the early outbreak in the U.S. and can greatly accelerate epidemics.
Keywords: SARS-CoV-2; genomic epidemiology; mobility; phylogenetics; viral emergence; viral sequencing.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.A.S. reports grants from the National Institutes of Health, European Research Council, and Wellcome Trust during the conduct of this research and grants and contracts from the Bill & Melinda Gates Foundation, Janssen Research and Development, Private Health Management, IQVIA, and the U.S. Department of Veterans Affairs outside the submitted work. S.L.L., R.R., and D.J.N. are employed by BioInfoexperts LLC. R.F.G. reports grants from the National Institutes of Health, the Coalition for Epidemic Preparedness Innovations, the Burroughs Wellcome Fund, the Wellcome Trust, the Center for Disease Prevention and Control, and the European & Developing Countries Clinical Trials Partnership. He is the co-founder and Chief Scientific Advisor of Zalgen Labs, a biotechnology company developing countermeasures to emerging viruses, including SARS-CoV-2. K.G.A. has received consulting fees and compensated expert testimony on SARS-CoV-2 and the COVID-19 pandemic.
Figures







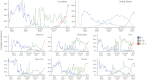

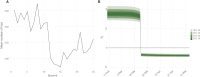
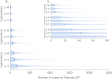
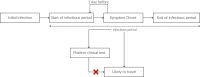
Update of
-
Emergence of an early SARS-CoV-2 epidemic in the United States.medRxiv [Preprint]. 2021 Feb 8:2021.02.05.21251235. doi: 10.1101/2021.02.05.21251235. medRxiv. 2021. Update in: Cell. 2021 Sep 16;184(19):4939-4952.e15. doi: 10.1016/j.cell.2021.07.030. PMID: 33564781 Free PMC article. Updated. Preprint.
References
-
- Ayres D.L., Cummings M.P., Baele G., Darling A.E., Lewis P.O., Swofford D.L., Huelsenbeck J.P., Lemey P., Rambaut A., Suchard M.A. BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2019;68:1052–1061. - PMC - PubMed
-
- Centers for Disease Control (2020a). https://www.cdc.gov/media/releases/2020/p0121-novel-coronavirus-travel-c... (Centers for Disease Control).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

