REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps
- PMID: 34508659
- PMCID: PMC8500949
- DOI: 10.1016/j.molcel.2021.08.016
REV1-Polζ maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps
Abstract
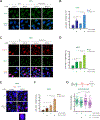
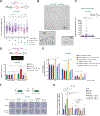
BRCA1/2 mutant tumor cells display an elevated mutation burden, the etiology of which remains unclear. Here, we report that these cells accumulate ssDNA gaps and spontaneous mutations during unperturbed DNA replication due to repriming by the DNA primase-polymerase PRIMPOL. Gap accumulation requires the DNA glycosylase SMUG1 and is exacerbated by depletion of the translesion synthesis (TLS) factor RAD18 or inhibition of the error-prone TLS polymerase complex REV1-Polζ by the small molecule JH-RE-06. JH-RE-06 treatment of BRCA1/2-deficient cells results in reduced mutation rates and PRIMPOL- and SMUG1-dependent loss of viability. Through cellular and animal studies, we demonstrate that JH-RE-06 is preferentially toxic toward HR-deficient cancer cells. Furthermore, JH-RE-06 remains effective toward PARP inhibitor (PARPi)-resistant BRCA1 mutant cells and displays additive toxicity with crosslinking agents or PARPi. Collectively, these studies identify a protective and mutagenic role for REV1-Polζ in BRCA1/2 mutant cells and provide the rationale for using REV1-Polζ inhibitors to treat BRCA1/2 mutant tumors.
Keywords: BRCA1 and BRCA2; DNA repriming; PRIMPOL; RAD18; REV1 and Polζ; breast and ovarian cancer; homologous recombination; ssDNA gaps; synthetic lethality; translesion synthesis.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

