Noncoding sequence variants define a novel regulatory element in the first intron of the N-acetylglutamate synthase gene
- PMID: 34510628
- PMCID: PMC8604755
- DOI: 10.1002/humu.24281
Noncoding sequence variants define a novel regulatory element in the first intron of the N-acetylglutamate synthase gene
Abstract
N-acetylglutamate synthase deficiency is an autosomal recessive urea cycle disorder caused either by decreased expression of the NAGS gene or defective NAGS enzyme resulting in decreased production of N-acetylglutamate (NAG), an allosteric activator of carbamylphosphate synthetase 1 (CPS1). NAGSD is the only urea cycle disorder that can be effectively treated with a single drug, N-carbamylglutamate (NCG), a stable NAG analog, which activates CPS1 to restore ureagenesis. We describe three patients with NAGSD due to four novel noncoding sequence variants in the NAGS regulatory regions. All three patients had hyperammonemia that resolved upon treatment with NCG. Sequence variants NM_153006.2:c.427-222G>A and NM_153006.2:c.427-218A>C reside in the 547 bp-long first intron of NAGS and define a novel NAGS regulatory element that binds retinoic X receptor α. Sequence variants NC_000017.10:g.42078967A>T (NM_153006.2:c.-3065A>T) and NC_000017.10:g.42078934C>T (NM_153006.2:c.-3098C>T) reside in the NAGS enhancer, within known HNF1 and predicted glucocorticoid receptor binding sites, respectively. Reporter gene assays in HepG2 and HuH-7 cells demonstrated that all four substitutions could result in reduced expression of NAGS. These findings show that analyzing noncoding regions of NAGS and other urea cycle genes can reveal molecular causes of disease and identify novel regulators of ureagenesis.
Keywords: N-acetylglutamate; N-acetylglutamate synthase; N-acetylglutamate synthase deficiency; intron; mutation analysis; noncoding sequence variants; regulatory element; urea cycle; urea cycle disorders.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
Conflicts of Interest
Mutation analysis (Dr. Häberle and Ms. Rüfenacht) for cases 2 and 3 and functional testing of the four non-coding variants (Dr. Caldovic and Ms. Haskins) were supported by the Recordati Rare Diseases, Inc. that manufactures and sells NCG as Carbaglu® (carglumic acid), which is used for treatment of
Figures

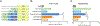

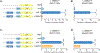
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

