Emerging Roles of LncRNAs in the EZH2-regulated Oncogenic Network
- PMID: 34512145
- PMCID: PMC8416728
- DOI: 10.7150/ijbs.63488
Emerging Roles of LncRNAs in the EZH2-regulated Oncogenic Network
Abstract
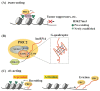
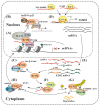
Cancer is a life-threatening disease, but cancer therapies based on epigenetic mechanisms have made great progress. Enhancer of zeste homolog 2 (EZH2) is the key catalytic component of Polycomb repressive complex 2 (PRC2) that mediates the tri-methylation of lysine 27 on histone 3 (H3K27me3), a well-recognized marker of transcriptional repression. Mounting evidence indicates that EZH2 is elevated in various cancers and associates with poor prognosis. In addition, many studies revealed that EZH2 is also involved in transcriptional repression dependent or independent of PRC2. Meanwhile, long non-coding RNAs (lncRNAs) have been reported to regulate numerous and diverse signaling pathways in oncogenesis. In this review, we firstly discuss functional interactions between EZH2 and lncRNAs that determine PRC2-dependent and -independent roles of EZH2. Secondly, we summarize the lncRNAs regulating EZH2 expression at transcription, post-transcription and post-translation levels. Thirdly, we review several oncogenic pathways cooperatively regulated by lncRNAs and EZH2, including the Wnt/β-catenin and p53 pathways. In conclusion, lncRNAs play a key role in the EZH2-regulated oncogenic network with many fertile directions to be explored.
Keywords: EZH2; H3K27me3; PRC2; cancer; epigenetic regulation; lncRNA; non-histone methylation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

