Discovering Potential Taxonomic Biomarkers of Type 2 Diabetes From Human Gut Microbiota via Different Feature Selection Methods
- PMID: 34512559
- PMCID: PMC8424122
- DOI: 10.3389/fmicb.2021.628426
Discovering Potential Taxonomic Biomarkers of Type 2 Diabetes From Human Gut Microbiota via Different Feature Selection Methods
Abstract
Human gut microbiota is a complex community of organisms including trillions of bacteria. While these microorganisms are considered as essential regulators of our immune system, some of them can cause several diseases. In recent years, next-generation sequencing technologies accelerated the discovery of human gut microbiota. In this respect, the use of machine learning techniques became popular to analyze disease-associated metagenomics datasets. Type 2 diabetes (T2D) is a chronic disease and affects millions of people around the world. Since the early diagnosis in T2D is important for effective treatment, there is an utmost need to develop a classification technique that can accelerate T2D diagnosis. In this study, using T2D-associated metagenomics data, we aim to develop a classification model to facilitate T2D diagnosis and to discover T2D-associated biomarkers. The sequencing data of T2D patients and healthy individuals were taken from a metagenome-wide association study and categorized into disease states. The sequencing reads were assigned to taxa, and the identified species are used to train and test our model. To deal with the high dimensionality of features, we applied robust feature selection algorithms such as Conditional Mutual Information Maximization, Maximum Relevance and Minimum Redundancy, Correlation Based Feature Selection, and select K best approach. To test the performance of the classification based on the features that are selected by different methods, we used random forest classifier with 100-fold Monte Carlo cross-validation. In our experiments, we observed that 15 commonly selected features have a considerable effect in terms of minimizing the microbiota used for the diagnosis of T2D and thus reducing the time and cost. When we perform biological validation of these identified species, we found that some of them are known as related to T2D development mechanisms and we identified additional species as potential biomarkers. Additionally, we attempted to find the subgroups of T2D patients using k-means clustering. In summary, this study utilizes several supervised and unsupervised machine learning algorithms to increase the diagnostic accuracy of T2D, investigates potential biomarkers of T2D, and finds out which subset of microbiota is more informative than other taxa by applying state-of-the art feature selection methods.
Keywords: classification; feature selection; human gut microbiome; machine learning; metagenomic analysis; type 2 diabetes.
Copyright © 2021 Bakir-Gungor, Bulut, Jabeer, Nalbantoglu and Yousef.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

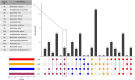

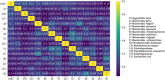


References
-
- Berthold M. R., Cebron N., Dill F., Gabriel T. R., Kötter T., Meinl T., et al. (2008). “KNIME: The Konstanz Information Miner,” in Data Analysis, Machine Learning and Applications. Studies in Classification, Data Analysis, and Knowledge Organization, eds Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R. (Berlin: Springer; ), 319–326. 10.1007/978-3-540-78246-9_38 - DOI
LinkOut - more resources
Full Text Sources

