Exome Sequencing Reveals a Putative Role for HLA-C*03:02 in Control of HIV-1 in African Pediatric Populations
- PMID: 34512729
- PMCID: PMC8428176
- DOI: 10.3389/fgene.2021.720213
Exome Sequencing Reveals a Putative Role for HLA-C*03:02 in Control of HIV-1 in African Pediatric Populations
Abstract
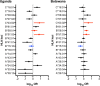
Human leucocyte antigen (HLA) class I molecules present endogenously processed antigens to T-cells and have been linked to differences in HIV-1 disease progression. HLA allelotypes show considerable geographical and inter-individual variation, as does the rate of progression of HIV-1 disease, with long-term non-progression (LTNP) of disease having most evidence of an underlying genetic contribution. However, most genetic analyses of LTNP have occurred in adults of European ancestry, limiting the potential transferability of observed associations to diverse populations who carry the burden of disease. This is particularly true of HIV-1 infected children. Here, using exome sequencing (ES) to infer HLA allelotypes, we determine associations with HIV-1 LTNP in two diverse African pediatric populations. We performed a case-control association study of 394 LTNPs and 420 rapid progressors retrospectively identified from electronic medical records of pediatric HIV-1 populations in Uganda and Botswana. We utilized high-depth ES to perform high-resolution HLA allelotyping and assessed evidence of association between HLA class I alleles and LTNP. Sixteen HLA alleles and haplotypes had significantly different frequencies between Uganda and Botswana, with allelic differences being more prominent in HLA-A compared to HLA-B and C allelotypes. Three HLA allelotypes showed association with LTNP, including a novel association in HLA-C (HLA-B∗57:03, aOR 3.21, Pc = 0.0259; B∗58:01, aOR 1.89, Pc = 0.033; C∗03:02, aOR 4.74, Pc = 0.033). Together, these alleles convey an estimated population attributable risk (PAR) of non-progression of 16.5%. We also observed novel haplotype associations with HLA-B∗57:03-C∗07:01 (aOR 5.40, Pc = 0.025) and HLA-B∗58:01-C∗03:02 (aOR 4.88, Pc = 0.011) with a PAR of 9.8%, as well as a previously unreported independent additive effect and heterozygote advantage of HLA-C∗03:02 with B∗58:01 (aOR 4.15, Pc = 0.005) that appears to limit disease progression, despite weak LD (r 2 = 0.18) between these alleles. These associations remained irrespective of gender or country. In one of the largest studies of HIV in Africa, we find evidence of a protective effect of canonical HLA-B alleles and a novel HLA-C association that appears to augment existing HIV-1 control alleles in pediatric populations. Our findings outline the value of using multi-ethnic populations in genetic studies and offer a novel HIV-1 association of relevance to ongoing vaccine studies.
Keywords: AIDS; childhood HIV; genetics; genomics; long-term non-progression.
Copyright © 2021 Kyobe, Mwesigwa, Kisitu, Farirai, Katagirya, Mirembe, Ketumile, Wayengera, Katabazi, Kigozi, Wampande, Retshabile, Mlotshwa, Williams, Morapedi, Kasvosve, Kyosiimire-Lugemwa, Nsangi, Tsimako-Johnstone, Brown, Joloba, Anabwani, Bhekumusa, Mpoloka, Mardon, Matshaba, Kekitiinwa and Hanchard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



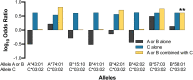
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

