Peptide-Based Inhibitors for SARS-CoV-2 and SARS-CoV
- PMID: 34514085
- PMCID: PMC8420164
- DOI: 10.1002/adtp.202100104
Peptide-Based Inhibitors for SARS-CoV-2 and SARS-CoV
Abstract
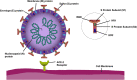
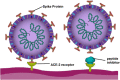
The COVID-19 (coronavirus disease) global pandemic, caused by the spread of the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) virus, currently has limited treatment options which include vaccines, anti-virals, and repurposed therapeutics. With their high specificity, tunability, and biocompatibility, small molecules like peptides are positioned to act as key players in combating SARS-CoV-2, and can be readily modified to match viral mutation rate. A recent expansion of the understanding of the viral structure and entry mechanisms has led to the proliferation of therapeutic viral entry inhibitors. In this comprehensive review, inhibitors of SARS and SARS-CoV-2 are investigated and discussed based on therapeutic design, inhibitory mechanistic approaches, and common targets. Peptide therapeutics are highlighted, which have demonstrated in vitro or in vivo efficacy, discuss advantages of peptide therapeutics, and common strategies in identifying targets for viral inhibition.
Keywords: SARS‐CoV; SARS‐CoV‐2; SARS‐CoV‐2 mutants; coronavirus; peptide therapeutics.
© 2021 Wiley‐VCH GmbH.
Conflict of interest statement
V.A.K. has equity interests in start‐up companies attempting to translate peptides from peptide‐based technological platform.
Figures


References
-
- Listings of WHO's response to COVID‐19, World Health Organization 2020, https://www.who.int/news/item/29-06-2020-covidtimeline.
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A., Berger A., Burguière A. M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J. C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H. D., Osterhaus A. D., Schmitz H., Doerr H. W., N. Engl. J. Med. 2003, 348, 1967. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
