RNA splicing programs define tissue compartments and cell types at single-cell resolution
- PMID: 34515025
- PMCID: PMC8563012
- DOI: 10.7554/eLife.70692
RNA splicing programs define tissue compartments and cell types at single-cell resolution
Abstract
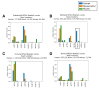
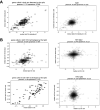
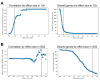
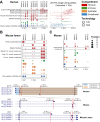
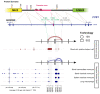
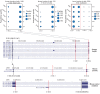
The extent splicing is regulated at single-cell resolution has remained controversial due to both available data and methods to interpret it. We apply the SpliZ, a new statistical approach, to detect cell-type-specific splicing in >110K cells from 12 human tissues. Using 10X Chromium data for discovery, 9.1% of genes with computable SpliZ scores are cell-type-specifically spliced, including ubiquitously expressed genes MYL6 and RPS24. These results are validated with RNA FISH, single-cell PCR, and Smart-seq2. SpliZ analysis reveals 170 genes with regulated splicing during human spermatogenesis, including examples conserved in mouse and mouse lemur. The SpliZ allows model-based identification of subpopulations indistinguishable based on gene expression, illustrated by subpopulation-specific splicing of classical monocytes involving an ultraconserved exon in SAT1. Together, this analysis of differential splicing across multiple organs establishes that splicing is regulated cell-type-specifically.
Keywords: RNA; computational biology; genetics; genomics; human; mouse; mouse lemur; scRNA-seq; splicing; statistics; systems biology.
© 2021, Olivieri et al.
Conflict of interest statement
JO, RD, PW, SJ, Ad, ST, JM, AR, SQ, MK, JS No competing interests declared
Figures


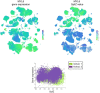
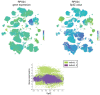
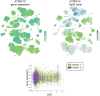


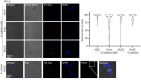






References
-
- Alexander D. STAR 2.7.9A. Github. 2021 https://github.com/alexdobin/STAR
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.14531721
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

