Fluorescence Imaging-Based Discovery of Membrane Domain-Associated Proteins in Mycobacterium smegmatis
- PMID: 34516286
- PMCID: PMC8544410
- DOI: 10.1128/JB.00419-21
Fluorescence Imaging-Based Discovery of Membrane Domain-Associated Proteins in Mycobacterium smegmatis
Abstract
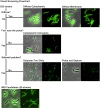
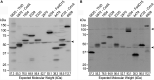
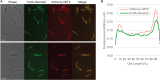
Mycobacteria spatially organize their plasma membrane, and many enzymes involved in envelope biosynthesis associate with a membrane compartment termed the intracellular membrane domain (IMD). The IMD is concentrated in the polar regions of growing cells and becomes less polarized under nongrowing conditions. Because mycobacteria elongate from the poles, the observed polar localization of the IMD during growth likely supports the localized biosynthesis of envelope components. While we have identified more than 300 IMD-associated proteins by proteomic analyses, only a few of these have been verified by independent experimental methods. Furthermore, some IMD-associated proteins may have escaped proteomic identification and remain to be identified. Here, we visually screened an arrayed library of 523 Mycobacterium smegmatis strains, each producing a Dendra2-FLAG-tagged recombinant protein. We identified 29 fusion proteins that showed polar fluorescence patterns characteristic of IMD proteins. Twenty of these had previously been suggested to localize to the IMD based on proteomic data. Of the nine remaining IMD candidate proteins, three were confirmed by biochemical methods to be associated with the IMD. Taken together, this new colocalization strategy is effective in verifying the IMD association of proteins found by proteomic analyses while facilitating the discovery of additional IMD-associated proteins. IMPORTANCE The intracellular membrane domain (IMD) is a membrane subcompartment found in Mycobacterium smegmatis cells. Proteomic analysis of purified IMD identified more than 300 proteins, including enzymes involved in cell envelope biosynthesis. However, proteomics on its own is unlikely to detect every IMD-associated protein because of technical and biological limitations. Here, we describe fluorescent protein colocalization as an alternative, independent approach. Using a combination of fluorescence microscopy, proteomics, and subcellular fractionation, we identified three new proteins associated with the IMD. Such a robust method to rigorously define IMD proteins will benefit future investigations to decipher the synthesis, maintenance, and functions of this membrane domain and help delineate a more general mechanism of subcellular protein localization in mycobacteria.
Keywords: Mycobacterial Systems Resource; fluorescence microscopy; membrane domain; membrane proteins; mycobacterium.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

