Nucleolar maturation of the human small subunit processome
- PMID: 34516797
- PMCID: PMC8744464
- DOI: 10.1126/science.abj5338
Nucleolar maturation of the human small subunit processome
Abstract
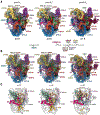
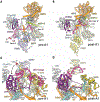
The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution cryo–electron microscopy structures of maturing human small subunit (SSU) processomes at resolutions of 2.7 to 3.9 angstroms. These structures reveal the molecular mechanisms that enable crucial progressions during SSU processome maturation. RNA folding states within these particles are communicated to and coordinated with key enzymes that drive irreversible steps such as targeted exosome-mediated RNA degradation, protein-guided site-specific endonucleolytic RNA cleavage, and tightly controlled RNA unwinding. These conserved mechanisms highlight the SSU processome’s impressive structural plasticity, which endows this 4.5-megadalton nucleolar assembly with the distinctive ability to mature the small ribosomal subunit from within.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

