Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination
- PMID: 34516917
- PMCID: PMC8442901
- DOI: 10.1126/sciadv.abj5365
Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination
Abstract
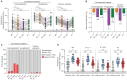
Emerging SARS-CoV-2 variants of concern (VOCs) pose a threat to human immunity induced by natural infection and vaccination. We assessed the recognition of three VOCs (B.1.1.7, B.1.351, and P.1) in cohorts of COVID-19 convalescent patients (n = 69) and Pfizer-BioNTech vaccine recipients (n = 50). Spike binding and neutralization against all three VOCs were substantially reduced in most individuals, with the largest four- to sevenfold reduction in neutralization being observed against B.1.351. While hospitalized patients with COVID-19 and vaccinees maintained sufficient neutralizing titers against all three VOCs, 39% of nonhospitalized patients exhibited no detectable neutralization against B.1.351. Moreover, monoclonal neutralizing antibodies show sharp reductions in their binding kinetics and neutralizing potential to B.1.351 and P.1 but not to B.1.1.7. These data have implications for the degree to which pre-existing immunity can protect against subsequent infection with VOCs and informs policy makers of susceptibility to globally circulating SARS-CoV-2 VOCs.
Figures


Update of
-
Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination.medRxiv [Preprint]. 2021 Jun 1:2021.05.26.21257441. doi: 10.1101/2021.05.26.21257441. medRxiv. 2021. Update in: Sci Adv. 2021 Sep 03;7(36):eabj5365. doi: 10.1126/sciadv.abj5365. PMID: 34100023 Free PMC article. Updated. Preprint.
References
-
- Weekly epidemiological update on COVID-19 - 13 April 2021; www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-1....
-
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D. L., Volz E.; COVID-19 Genomics Consortium UK (CoG-UK) , Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Genom. Epidemiol. , (2020).
-
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E. J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A. J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.-Y., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C. K., Sewell B. T., Lourenço J., Alcantara L. C. J., Pond S. L. K., Weaver S., Martin D., Lessells R. J., Bhiman J. N., Williamson C., de Oliveira T., Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443 (2021). - PubMed
-
- N. R. Faria, I. M. Claro, D. Candido, L. A. Moyses Franco, P. S. Andrade, T. M. Coletti, C. A. M. Silva, F. C. Sales, E. R. Manuli, R. S. Aguiar, Others, “Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings” (2021); www.icpcovid.com/sites/default/files/2021-01/Ep%20102-1%20Genomic%20char....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

