Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes
- PMID: 34516923
- PMCID: PMC8442896
- DOI: 10.1126/sciadv.abe8215
Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes
Abstract
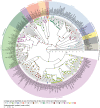
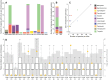
Sex is a fundamental trait determined by environmental and/or genetic factors, including sex chromosomes. Sex chromosomes are studied in species scattered across the tree of life, yet little is known about tempo and mode of sex chromosome evolution among closely related species. Here, we examine sex chromosome evolution in the adaptive radiation of cichlid fishes in Lake Tanganyika. Through the analysis of male and female genomes from 244 cichlid taxa (189 described species with 5 represented with two local variants/populations; 50 undescribed species) and of 396 multitissue transcriptomes from 66 taxa, we identify signatures of sex chromosomes in 79 taxa, involving 12 linkage groups. We find that Tanganyikan cichlids have the highest rates of sex chromosome turnover and heterogamety transitions known to date. We show that sex chromosome recruitment is not at random. Moreover convergently emerged sex chromosomes in cichlids support the “limited options” hypothesis of sex chromosome evolution.
Figures



References
-
- Rice W. R., Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742 (1984). - PubMed
-
- Charlesworth B., Coyne J. A., Barton N. H., The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130, 113–146 (1987).
-
- Bull J. J., Charnov E. L., Changes in the heterogametic mechanism of sex determination. Heredity 39, 1–14 (1977). - PubMed
-
- Blaser O., Grossen C., Neuenschwander S., Perrin N., Sex-chromosome turnovers induced by deleterious mutation load. Evolution 67, 635–645 (2013). - PubMed
LinkOut - more resources
Full Text Sources

