TnCentral: a Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis
- PMID: 34517763
- PMCID: PMC8546635
- DOI: 10.1128/mBio.02060-21
TnCentral: a Prokaryotic Transposable Element Database and Web Portal for Transposon Analysis
Abstract
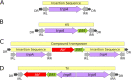
We describe here the structure and organization of TnCentral (https://tncentral.proteininformationresource.org/ [or the mirror link at https://tncentral.ncc.unesp.br/]), a web resource for prokaryotic transposable elements (TE). TnCentral currently contains ∼400 carefully annotated TE, including transposons from the Tn3, Tn7, Tn402, and Tn554 families; compound transposons; integrons; and associated insertion sequences (IS). These TE carry passenger genes, including genes conferring resistance to over 25 classes of antibiotics and nine types of heavy metal, as well as genes responsible for pathogenesis in plants, toxin/antitoxin gene pairs, transcription factors, and genes involved in metabolism. Each TE has its own entry page, providing details about its transposition genes, passenger genes, and other sequence features required for transposition, as well as a graphical map of all features. TnCentral content can be browsed and queried through text- and sequence-based searches with a graphic output. We describe three use cases, which illustrate how the search interface, results tables, and entry pages can be used to explore and compare TE. TnCentral also includes downloadable software to facilitate user-driven identification, with manual annotation, of certain types of TE in genomic sequences. Through the TnCentral homepage, users can also access TnPedia, which provides comprehensive reviews of the major TE families, including an extensive general section and specialized sections with descriptions of insertion sequence and transposon families. TnCentral and TnPedia are intuitive resources that can be used by clinicians and scientists to assess TE diversity in clinical, veterinary, and environmental samples. IMPORTANCE The ability of bacteria to undergo rapid evolution and adapt to changing environmental circumstances drives the public health crisis of multiple antibiotic resistance, as well as outbreaks of disease in economically important agricultural crops and animal husbandry. Prokaryotic transposable elements (TE) play a critical role in this. Many carry "passenger genes" (not required for the transposition process) conferring resistance to antibiotics or heavy metals or causing disease in plants and animals. Passenger genes are spread by normal TE transposition activities and by insertion into plasmids, which then spread via conjugation within and across bacterial populations. Thus, an understanding of TE composition and transposition mechanisms is key to developing strategies to combat bacterial pathogenesis. Toward this end, we have developed TnCentral, a bioinformatics resource dedicated to describing and exploring the structural and functional features of prokaryotic TE whose use is intuitive and accessible to users with or without bioinformatics expertise.
Keywords: antibiotic resistance; database; genome evolution; insertion sequence; integrons; mobile genetic elements; plasmid-mediated resistance; transposition mechanisms; transposons; virulence.
Figures





References
-
- Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J, Infectious Diseases Society of America . 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46:155–164. doi:10.1086/524891. - DOI - PubMed
-
- O’Neill J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Welcome Foundation HM Government UK, London, United Kingdom.
-
- Craig NL (ed). 2015. Mobile DNA III. American Society of Microbiology, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

