Oxidative Phosphorylation Is a Metabolic Vulnerability in Chemotherapy-Resistant Triple-Negative Breast Cancer
- PMID: 34518211
- PMCID: PMC8563442
- DOI: 10.1158/0008-5472.CAN-20-3242
Oxidative Phosphorylation Is a Metabolic Vulnerability in Chemotherapy-Resistant Triple-Negative Breast Cancer
Abstract
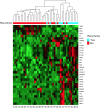
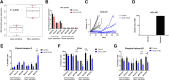
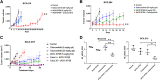
Oxidative phosphorylation (OXPHOS) is an active metabolic pathway in many cancers. RNA from pretreatment biopsies from patients with triple-negative breast cancer (TNBC) who received neoadjuvant chemotherapy demonstrated that the top canonical pathway associated with worse outcome was higher expression of OXPHOS signature. IACS-10759, a novel inhibitor of OXPHOS, stabilized growth in multiple TNBC patient-derived xenografts (PDX). On gene expression profiling, all of the sensitive models displayed a basal-like 1 TNBC subtype. Expression of mitochondrial genes was significantly higher in sensitive PDXs. An in vivo functional genomics screen to identify synthetic lethal targets in tumors treated with IACS-10759 found several potential targets, including CDK4. We validated the antitumor efficacy of the combination of palbociclib, a CDK4/6 inhibitor, and IACS-10759 in vitro and in vivo. In addition, the combination of IACS-10759 and multikinase inhibitor cabozantinib had improved antitumor efficacy. Taken together, our data suggest that OXPHOS is a metabolic vulnerability in TNBC that may be leveraged with novel therapeutics in combination regimens. SIGNIFICANCE: These findings suggest that triple-negative breast cancer is highly reliant on OXPHOS and that inhibiting OXPHOS may be a novel approach to enhance efficacy of several targeted therapies.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures



References
-
- Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

