TRIM28 is a transcriptional activator of the mutant TERT promoter in human bladder cancer
- PMID: 34518220
- PMCID: PMC8463889
- DOI: 10.1073/pnas.2102423118
TRIM28 is a transcriptional activator of the mutant TERT promoter in human bladder cancer
Abstract
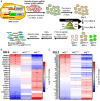
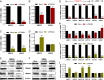
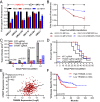
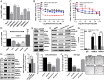
Bladder cancer (BC) has a 70% telomerase reverse transcriptase (TERT or hTERT in humans) promoter mutation prevalence, commonly at -124 base pairs, and this is associated with increased hTERT expression and poor patient prognosis. We inserted a green fluorescent protein (GFP) tag in the mutant hTERT promoter allele to create BC cells expressing an hTERT-GFP fusion protein. These cells were used in a fluorescence-activated cell sorting-based pooled CRISPR-Cas9 Kinome knockout genetic screen to identify tripartite motif containing 28 (TRIM28) and TRIM24 as regulators of hTERT expression. TRIM28 activates, while TRIM24 suppresses, hTERT transcription from the mutated promoter allele. TRIM28 is recruited to the mutant promoter where it interacts with TRIM24, which inhibits its activity. Phosphorylation of TRIM28 through the mTOR complex 1 (mTORC1) releases it from TRIM24 and induces hTERT transcription. TRIM28 expression promotes in vitro and in vivo BC cell growth and stratifies BC patient outcome. mTORC1 inhibition with rapamycin analog Ridaforolimus suppresses TRIM28 phosphorylation, hTERT expression, and cell viability. This study may lead to hTERT-directed cancer therapies with reduced effects on normal progenitor cells.
Keywords: CRISPR-Cas9 KnockIn; Kinome KO screening; hTERT; promoter mutation.
Conflict of interest statement
Competing interest statement: T.R.C. is on the board of directors of Merck, Inc. and a scientific advisor to Storm Therapeutics and Eikon Therapeutics.
Figures



Comment in
-
Uro-Science.J Urol. 2022 Jun;207(6):1341-1342. doi: 10.1097/JU.0000000000002656. Epub 2022 Mar 18. J Urol. 2022. PMID: 35300509 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

