The taxonomy of two uncultivated fungal mammalian pathogens is revealed through phylogeny and population genetic analyses
- PMID: 34518564
- PMCID: PMC8438014
- DOI: 10.1038/s41598-021-97429-7
The taxonomy of two uncultivated fungal mammalian pathogens is revealed through phylogeny and population genetic analyses
Abstract
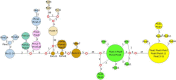
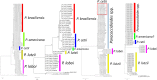
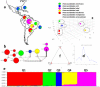
Ever since the uncultivated South American fungal pathogen Lacazia loboi was first described 90 years ago, its etiology and evolutionary traits have been at the center of endless controversies. This pathogen infects the skin of humans and as long believed, dolphin skin. However, recent DNA analyses of infected dolphins placed its DNA sequences within Paracoccidioides species. This came as a surprise and suggested the human and dolphin pathogens may be different species. In this study, population genetic analyses of DNA from four infected dolphins grouped this pathogen in a monophyletic cluster sister to P. americana and to the other Paracoccidioides species. Based on the results we have emended the taxonomy of the dolphin pathogen as Paracoccidioides cetii and P. loboi the one infecting human. Our data warn that phylogenetic analysis of available taxa without the inclusion of unusual members may provide incomplete information for the accurate classification of anomalous species.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lôbo J. Um caso de blastomycose produzido por uma espécie nova encontrada em Recife. Rev. Med. Perenambuco. 1931;1:763–765.
-
- Lacaz CS, Baruzzi RG, Rosa MCB. Doença de Jorge Lôbo. IPSIS Gráfica e Editora; 1986. pp. 1–92.
-
- Baruzzi RG, Lacaz S, Souza PPA. História natural da doença de Jorge Lôbo. Occurrência entre os índios caiabi (Brasil central) Rev. Med. Trop. São Paulo. 1979;21:302–338. - PubMed
-
- Talhari S, Cunha MGS, Barros MLB, Gadelha AR. Doença de Jorge Lôbo. Estudo de 22 casos novos. Med. Cut. Ibero. Latin Amer. 1981;9:87–96. - PubMed
-
- Migaki G, Valerio MG, Irvine B, Garner FM. Lobo’s disease in an Atlantic bottle nosed dolphin. J. Am. Vet. Med. Assoc. 1971;159:578–582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

