A regulatory cascade controls Staphylococcus aureus pathogenicity island activation
- PMID: 34518655
- PMCID: PMC7611864
- DOI: 10.1038/s41564-021-00956-2
A regulatory cascade controls Staphylococcus aureus pathogenicity island activation
Abstract
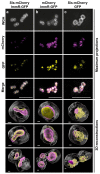
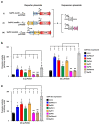
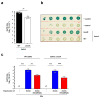
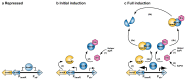
Staphylococcal pathogenicity islands (SaPIs) are a family of closely related mobile chromosomal islands that encode and disseminate the superantigen toxins, toxic shock syndrome toxin 1 and superantigen enterotoxin B (SEB). They are regulated by master repressors, which are counteracted by helper phage-encoded proteins, thereby inducing their excision, replication, packaging and intercell transfer. SaPIs are major components of the staphylococcal mobilome, occupying five chromosomal att sites, with many strains harbouring two or more. As regulatory interactions between co-resident SaPIs could have profound effects on the spread of superantigen pathobiology, we initiated the current study to search for such interactions. Using classical genetics, we found that, with one exception, their regulatory systems do not cross-react. The exception was SaPI3, which was originally considered defective because it could not be mobilized by any known helper phage. We show here that SaPI3 has an atypical regulatory module and is induced not by a phage but by many other SaPIs, including SaPI2, SaPIbov1 and SaPIn1, each encoding a conserved protein, Sis, which counteracts the SaPI3 repressor, generating an intracellular regulatory cascade: the co-resident SaPI, when conventionally induced by a helper phage, expresses its sis gene which, in turn, induces SaPI3, enabling it to spread. Using bioinformatics analysis, we have identified more than 30 closely related coancestral SEB-encoding SaPI3 relatives occupying the same att site and controlled by a conserved regulatory module, immA-immR-str'. This module is functionally analogous but unrelated to the typical SaPI regulatory module, stl-str. As SaPIs are phage satellites, SaPI3 and its relatives are SaPI satellites.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures


References
-
- Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus . Mol Microbiol. 1998;29:527–543. - PubMed
-
- Gillet Y, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. - PubMed
-
- Penadés JR, Christie GE. The phage-inducible chromosomal islands: a family of highly evolved molecular parasites. Annu Rev Virol. 2015;2:181–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

