Formation of a protein corona on the surface of extracellular vesicles in blood plasma
- PMID: 34520123
- PMCID: PMC8439280
- DOI: 10.1002/jev2.12140
Formation of a protein corona on the surface of extracellular vesicles in blood plasma
Abstract
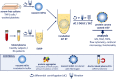
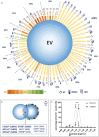
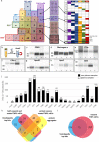
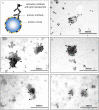
In this study we tested whether a protein corona is formed around extracellular vesicles (EVs) in blood plasma. We isolated medium-sized nascent EVs of THP1 cells as well as of Optiprep-purified platelets, and incubated them in EV-depleted blood plasma from healthy subjects and from patients with rheumatoid arthritis. EVs were subjected to differential centrifugation, size exclusion chromatography, or density gradient ultracentrifugation followed by mass spectrometry. Plasma protein-coated EVs had a higher density compared to the nascent ones and carried numerous newly associated proteins. Interactions between plasma proteins and EVs were confirmed by confocal microscopy, capillary Western immunoassay, immune electron microscopy and flow cytometry. We identified nine shared EV corona proteins (ApoA1, ApoB, ApoC3, ApoE, complement factors 3 and 4B, fibrinogen α-chain, immunoglobulin heavy constant γ2 and γ4 chains), which appear to be common corona proteins among EVs, viruses and artificial nanoparticles in blood plasma. An unexpected finding of this study was the high overlap of the composition of the protein corona with blood plasma protein aggregates. This is explained by our finding that besides a diffuse, patchy protein corona, large protein aggregates also associate with the surface of EVs. However, while EVs with an external plasma protein cargo induced an increased expression of TNF-α, IL-6, CD83, CD86 and HLA-DR of human monocyte-derived dendritic cells, EV-free protein aggregates had no effect. In conclusion, our data may shed new light on the origin of the commonly reported plasma protein 'contamination' of EV preparations and may add a new perspective to EV research.
Keywords: aggregation; blood plasma; extracellular vesicles; mass spectrometry; protein corona.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
EIB is member of the Advisory Board of Sphere Gene Therapeutics Inc. (Boston, USA).
Figures




References
-
- Aletaha, D., Neogi, T., Silman, A J., Funovits, J., Felson, D T., Bingham, C O., Birnbaum, N S., Burmester, G R., Bykerk, V P., Cohen, M D., Combe, B., Costenbader, K H., Dougados, M., Emery, P., Ferraccioli, G., Hazes, J M. W., Hobbs, K., Huizinga, T W. J., Kavanaugh, A.…Hawker, G. (2010). Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis and Rheumatism, 62(9), 2569–2581. 10.1002/art.27584 - DOI - PubMed
-
- Bateman, A., Martin, M. J., Orchard, S., Magrane, M., Agivetova, R., Ahmad, S., Alpi, E., Bowler‐Barnett, E. H., Britto, R., Bursteinas, B., Bye‐A‐Jee, H., Coetzee, R., Cukura, A., Da Silva, A., Denny, P., Dogan, T., Ebenezer, T., Fan, J., Castro, L. G.…Teodoro, D. (2021).UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research, 49(D1), D480–D489. 10.1093/nar/gkaa1100 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

