Novel strategy for disease risk prediction incorporating predicted gene expression and DNA methylation data: a multi-phased study of prostate cancer
- PMID: 34520132
- PMCID: PMC8696216
- DOI: 10.1002/cac2.12205
Novel strategy for disease risk prediction incorporating predicted gene expression and DNA methylation data: a multi-phased study of prostate cancer
Abstract
Background: DNA methylation and gene expression are known to play important roles in the etiology of human diseases such as prostate cancer (PCa). However, it has not yet been possible to incorporate information of DNA methylation and gene expression into polygenic risk scores (PRSs). Here, we aimed to develop and validate an improved PRS for PCa risk by incorporating genetically predicted gene expression and DNA methylation, and other genomic information using an integrative method.
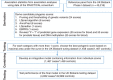
Methods: Using data from the PRACTICAL consortium, we derived multiple sets of genetic scores, including those based on available single-nucleotide polymorphisms through widely used methods of pruning and thresholding, LDpred, LDpred-funt, AnnoPred, and EBPRS, as well as PRS constructed using the genetically predicted gene expression and DNA methylation through a revised pruning and thresholding strategy. In the tuning step, using the UK Biobank data (1458 prevalent cases and 1467 controls), we selected PRSs with the best performance. Using an independent set of data from the UK Biobank, we developed an integrative PRS combining information from individual scores. Furthermore, in the testing step, we tested the performance of the integrative PRS in another independent set of UK Biobank data of incident cases and controls.
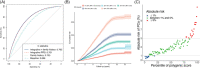
Results: Our constructed PRS had improved performance (C statistics: 76.1%) over PRSs constructed by individual benchmark methods (from 69.6% to 74.7%). Furthermore, our new PRS had much higher risk assessment power than family history. The overall net reclassification improvement was 69.0% by adding PRS to the baseline model compared with 12.5% by adding family history.
Conclusions: We developed and validated a new PRS which may improve the utility in predicting the risk of developing PCa. Our innovative method can also be applied to other human diseases to improve risk prediction across multiple outcomes.
Keywords: integrative models; polygenic risk scores; predicted DNA methylation; predicted gene expression; prostate cancer; risk prediction.
© 2021 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
No potential conflicts of interest were disclosed by the authors.
Figures
References
Publication types
MeSH terms
Grants and funding
- 251533/The National Health and Medical Research Council, Australia
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- U01 CA188392/CA/NCI NIH HHS/United States
- U01 CA261339/CA/NCI NIH HHS/United States
- R03 AG070669/AG/NIA NIH HHS/United States
- CIHR/Canada
- U19 CA148537/CA/NCI NIH HHS/United States
- X01HG007492/Prostate cancer SuscEptibility (ELLIPSE)
- R01 CA128813/CA/NCI NIH HHS/United States
- 12-823/Swedish Cancer Foundation
- 940394/The National Health and Medical Research Council, Australia
- 2014-2269/Swedish Research Council, Swedish Research Council
- R03 AG070669/GF/NIH HHS/United States
- HEALTH-F2-2009-223175/Canadian Institutes of Health Research, European Commission's Seventh Framework Programme grant agreement
- 126402/The National Health and Medical Research Council, Australia
- 11-484/Swedish Cancer Foundation
- U01-CA98758/U.S. National Institutes of Health, National Cancer Institute
- 396414/The National Health and Medical Research Council, Australia
- K2010-70X-20430-04-3/Swedish Research Council, Swedish Research Council
- C5047/A10692/CRUK_/Cancer Research UK/United Kingdom
- U01 CA098216/CA/NCI NIH HHS/United States
- HHSN268201200008C/HL/NHLBI NIH HHS/United States
- C5047/A7357/CRUK_/Cancer Research UK/United Kingdom
- R01 CA128978/CA/NCI NIH HHS/United States
- U01-CA98710/U.S. National Institutes of Health, National Cancer Institute
- C1287/A10118/CRUK_/Cancer Research UK/United Kingdom
- 1 U19 CA 148537-01/The National Institute of Health (NIH) Cancer Post-Cancer GWAS
- 504700/The National Health and Medical Research Council, Australia
- 504702/The National Health and Medical Research Council, Australia
- U01 CA098233/CA/NCI NIH HHS/United States
- U01-CA98233/U.S. National Institutes of Health, National Cancer Institute
- C16913/A6135/CRUK_/Cancer Research UK/United Kingdom
- 504715/The National Health and Medical Research Council, Australia
- 09-0677/Swedish Cancer Foundation
- 1U19 CA148112/Post-Cancer GWAS initiative
- U19 CA148112/CA/NCI NIH HHS/United States
- U01-CA98216/U.S. National Institutes of Health, National Cancer Institute
- U01 CA098758/CA/NCI NIH HHS/United States
- U19 CA 148537/US National Institutes of Health (NIH)
- 623204/The National Health and Medical Research Council, Australia
- U19 CA148065/CA/NCI NIH HHS/United States
- C5047/A3354/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 223175/European Community's Seventh Framework Programme
- HHSN268201200008I/HL/NHLBI NIH HHS/United States
- 614296/The National Health and Medical Research Council, Australia
- P30 CA071789/CA/NCI NIH HHS/United States
- C1287/A16563/CRUK_/Cancer Research UK/United Kingdom
- 1U19 CA148065/Post-Cancer GWAS initiative
- 209057/WT_/Wellcome Trust/United Kingdom
- U01 CA098710/CA/NCI NIH HHS/United States
- 450104/The National Health and Medical Research Council, Australia
- 1U19 CA148537/Post-Cancer GWAS initiative
LinkOut - more resources
Full Text Sources
Medical

