Joint effects of genes underlying a temperature specialization tradeoff in yeast
- PMID: 34520469
- PMCID: PMC8462698
- DOI: 10.1371/journal.pgen.1009793
Joint effects of genes underlying a temperature specialization tradeoff in yeast
Abstract
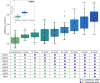
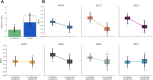
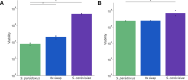
A central goal of evolutionary genetics is to understand, at the molecular level, how organisms adapt to their environments. For a given trait, the answer often involves the acquisition of variants at unlinked sites across the genome. Genomic methods have achieved landmark successes in pinpointing these adaptive loci. To figure out how a suite of adaptive alleles work together, and to what extent they can reconstitute the phenotype of interest, requires their transfer into an exogenous background. We studied the joint effect of adaptive, gain-of-function thermotolerance alleles at eight unlinked genes from Saccharomyces cerevisiae, when introduced into a thermosensitive sister species, S. paradoxus. Although the loci damped each other's beneficial impact (that is, they were subject to negative epistasis), most boosted high-temperature growth alone and in combination, and none was deleterious. The complete set of eight genes was sufficient to confer ~15% of the S. cerevisiae thermotolerance phenotype in the S. paradoxus background. The same loci also contributed to a heretofore unknown advantage in cold growth by S. paradoxus. Together, our data establish temperature resistance in yeasts as a model case of a genetically complex evolutionary tradeoff, which can be partly reconstituted from the sequential assembly of unlinked underlying loci.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

