Theoretical basis for stabilizing messenger RNA through secondary structure design
- PMID: 34520542
- PMCID: PMC8499941
- DOI: 10.1093/nar/gkab764
Theoretical basis for stabilizing messenger RNA through secondary structure design
Erratum in
-
Correction to 'Theoretical basis for stabilizing messenger RNA through secondary structure design'.Nucleic Acids Res. 2021 Nov 8;49(19):11405. doi: 10.1093/nar/gkab911. Nucleic Acids Res. 2021. PMID: 34591967 Free PMC article. No abstract available.
Abstract
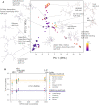
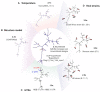
RNA hydrolysis presents problems in manufacturing, long-term storage, world-wide delivery and in vivo stability of messenger RNA (mRNA)-based vaccines and therapeutics. A largely unexplored strategy to reduce mRNA hydrolysis is to redesign RNAs to form double-stranded regions, which are protected from in-line cleavage and enzymatic degradation, while coding for the same proteins. The amount of stabilization that this strategy can deliver and the most effective algorithmic approach to achieve stabilization remain poorly understood. Here, we present simple calculations for estimating RNA stability against hydrolysis, and a model that links the average unpaired probability of an mRNA, or AUP, to its overall hydrolysis rate. To characterize the stabilization achievable through structure design, we compare AUP optimization by conventional mRNA design methods to results from more computationally sophisticated algorithms and crowdsourcing through the OpenVaccine challenge on the Eterna platform. We find that rational design on Eterna and the more sophisticated algorithms lead to constructs with low AUP, which we term 'superfolder' mRNAs. These designs exhibit a wide diversity of sequence and structure features that may be desirable for translation, biophysical size, and immunogenicity. Furthermore, their folding is robust to temperature, computer modeling method, choice of flanking untranslated regions, and changes in target protein sequence, as illustrated by rapid redesign of superfolder mRNAs for B.1.351, P.1 and B.1.1.7 variants of the prefusion-stabilized SARS-CoV-2 spike protein. Increases in in vitro mRNA half-life by at least two-fold appear immediately achievable.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Update of
-
Theoretical basis for stabilizing messenger RNA through secondary structure design.bioRxiv [Preprint]. 2021 Feb 19:2020.08.22.262931. doi: 10.1101/2020.08.22.262931. bioRxiv. 2021. Update in: Nucleic Acids Res. 2021 Oct 11;49(18):10604-10617. doi: 10.1093/nar/gkab764. PMID: 32869022 Free PMC article. Updated. Preprint.
References
-
- Chauhan G., Madou M.J., Kalra S., Chopra V., Ghosh D., Martinez-Chapa S.O.. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020; 14:7760–7782. - PubMed
-
- Verbeke R., Lentacker I., De Smedt S.C., Dewitte H.. Three decades of messenger RNA vaccine development. Nano Today. 2019; 28:100766.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

