PathFams: statistical detection of pathogen-associated protein domains
- PMID: 34521345
- PMCID: PMC8442362
- DOI: 10.1186/s12864-021-07982-8
PathFams: statistical detection of pathogen-associated protein domains
Abstract
Background: A substantial fraction of genes identified within bacterial genomes encode proteins of unknown function. Identifying which of these proteins represent potential virulence factors, and mapping their key virulence determinants, is a challenging but important goal.
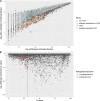
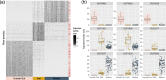
Results: To facilitate virulence factor discovery, we performed a comprehensive analysis of 17,929 protein domain families within the Pfam database, and scored them based on their overrepresentation in pathogenic versus non-pathogenic species, taxonomic distribution, relative abundance in metagenomic datasets, and other factors.
Conclusions: We identify pathogen-associated domain families, candidate virulence factors in the human gut, and eukaryotic-like mimicry domains with likely roles in virulence. Furthermore, we provide an interactive database called PathFams to allow users to explore pathogen-associated domains as well as identify pathogen-associated domains and domain architectures in user-uploaded sequences of interest. PathFams is freely available at https://pathfams.uwaterloo.ca .
Keywords: Environmental association; Hypothetical proteins; Lineage specificity; Pathogens; Proteins of unknown function; Virulence factors.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

