Acid resistance system CadBA is implicated in acid tolerance and biofilm formation and is identified as a new virulence factor of Edwardsiella tarda
- PMID: 34521475
- PMCID: PMC8438976
- DOI: 10.1186/s13567-021-00987-x
Acid resistance system CadBA is implicated in acid tolerance and biofilm formation and is identified as a new virulence factor of Edwardsiella tarda
Abstract
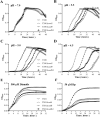
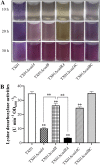
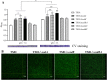
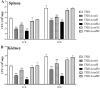
Edwardsiella tarda is a facultative intracellular pathogen in humans and animals. The Gram-negative bacterium is widely considered a potentially important bacterial pathogen. Adaptation to acid stress is important for the transmission of intestinal microbes, so the acid-resistance (AR) system is essential. However, the AR systems of E. tarda are totally unknown. In this study, a lysine-dependent acid resistance (LDAR) system in E. tarda, CadBA, was characterized and identified. CadB is a membrane protein and shares high homology with the lysine/cadaverine antiporter. CadA contains a PLP-binding core domain and a pyridoxal phosphate-binding motif. It shares high homology with lysine decarboxylase. cadB and cadA are co-transcribed under one operon. To study the function of the cadBA operon, isogenic cadA, cadB and cadBA deletion mutant strains TX01ΔcadA, TX01ΔcadB and TX01ΔcadBA were constructed. When cultured under normal conditions, the wild type strain and three mutants exhibited the same growth performance. However, when cultured under acid conditions, the growth of three mutants, especially TX01ΔcadA, were obviously retarded, compared to the wild strain TX01, which indicates the important involvement of the cadBA operon in acid resistance. The deletion of cadB or cadA, especially cadBA, significantly attenuated bacterial activity of lysine decarboxylase, suggesting the vital participation of cadBA operon in lysine metabolism, which is closely related to acid resistance. The mutations of cadBA operon enhanced bacterial biofilm formation, especially under acid conditions. The deletions of the cadBA operon reduced bacterial adhesion and invasion to Hela cells. Consistently, the deficiency of cadBA operon abated bacterial survival and replication in macrophages, and decreased bacterial dissemination in fish tissues. Our results also show that the expression of cadBA operon and regulator cadC were up-regulated upon acid stress, and CadC rigorously regulated the expression of cadBA operon, especially under acid conditions. These findings demonstrate that the AR CadBA system was a requisite for the resistance of E. tarda against acid stress, and played a critical role in bacterial infection of host cells and in host tissues. This is the first study about the acid resistance system of E. tarda and provides new insights into the acid-resistance mechanism and pathogenesis of E. tarda.
Keywords: Edwardsiella tarda; acid resistance; biofilm; cadBA; pathogenicity; regulation.
© 2021. The Author(s).
Conflict of interest statement
The authors have no conflicting commercial or financial interest in publishing this paper.
Figures





References
-
- Ewing WH, Mcwhorter AC, Escobar MR, Lubin AH. Edwardsiella, a new genus of Enterobacteriaceae based on a new species, E. tarda. Int J Syst Evol Microbiol. 1965;15:33–38.
-
- Griffin MJ, Quiniou SM, Cody T, Tabuchi M, Ware C, Cipriano RC, Mauel MJ, Soto E. Comparative analysis of Edwardsiella isolates from fish in the eastern United States identifies two distinct genetic taxa amongst organisms phenotypically classified as E. tarda. Vet Microbiol. 2013;165:358–372. doi: 10.1016/j.vetmic.2013.03.027. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

