New-onset IgG autoantibodies in hospitalized patients with COVID-19
- PMID: 34521836
- PMCID: PMC8440763
- DOI: 10.1038/s41467-021-25509-3
New-onset IgG autoantibodies in hospitalized patients with COVID-19
Abstract
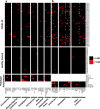
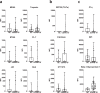
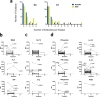
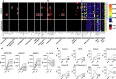
COVID-19 is associated with a wide range of clinical manifestations, including autoimmune features and autoantibody production. Here we develop three protein arrays to measure IgG autoantibodies associated with connective tissue diseases, anti-cytokine antibodies, and anti-viral antibody responses in serum from 147 hospitalized COVID-19 patients. Autoantibodies are identified in approximately 50% of patients but in less than 15% of healthy controls. When present, autoantibodies largely target autoantigens associated with rare disorders such as myositis, systemic sclerosis and overlap syndromes. A subset of autoantibodies targeting traditional autoantigens or cytokines develop de novo following SARS-CoV-2 infection. Autoantibodies track with longitudinal development of IgG antibodies recognizing SARS-CoV-2 structural proteins and a subset of non-structural proteins, but not proteins from influenza, seasonal coronaviruses or other pathogenic viruses. We conclude that SARS-CoV-2 causes development of new-onset IgG autoantibodies in a significant proportion of hospitalized COVID-19 patients and are positively correlated with immune responses to SARS-CoV-2 proteins.
© 2021. The Author(s).
Conflict of interest statement
C.S. received consultancy fees and research funding from Hycor Biomedical and Thermo Fisher Scientific, research funding from Mead Johnson Nutrition (MJN), and consultancy fees from Bencard Allergie. E.J.W. has consulting agreements with and/or is on the scientific advisory board for Merck, Elstar, Janssen, Related Sciences, Synthekine, and Surface Oncology. E.J.W. is a founder of Surface Oncology and Arsenal Biosciences. E.J.W. has a patent licensing agreement on the PD-1 pathway with Roche/Genentech. E.M. received consultancy fees from Roche. E.T.L.P. receives research funding from Janssen Research and Development, consultancy fees, and research funding from Roche Diagnostics, is a paid consultant for Enpicom, and serves on the scientific advisory boards of the Antibody Society, the Immune Epitope Database, and the American Autoimmune Related Diseases Association. N.J.M. reports funding to her institution from Athersys Inc, Biomarck Inc, and Quantum Leap Healthcare Collaborative, outside of the funded work. S. Chinthrajah reports grants from NIAID, CoFAR, Aimmune, DBV Technologies, Astellas, Regeneron, FARE, and is an Advisory Board member for Alladapt, Genentech, Novartis, Sanofi, and received personal fees from Nutricia. All remaining authors declare no competing interests with the research reported in this paper.
Figures

Update of
-
New-Onset IgG Autoantibodies in Hospitalized Patients with COVID-19.medRxiv [Preprint]. 2021 Jan 29:2021.01.27.21250559. doi: 10.1101/2021.01.27.21250559. medRxiv. 2021. Update in: Nat Commun. 2021 Sep 14;12(1):5417. doi: 10.1038/s41467-021-25509-3. PMID: 33532787 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- L40 AI120105/AI/NIAID NIH HHS/United States
- R01 AI125197/AI/NIAID NIH HHS/United States
- UM1 AI144292/AI/NIAID NIH HHS/United States
- U19 AI117950/AI/NIAID NIH HHS/United States
- U19 AI082630/AI/NIAID NIH HHS/United States
- P01 AI112521/AI/NIAID NIH HHS/United States
- P30 CA016520/CA/NCI NIH HHS/United States
- UC4 DK112217/DK/NIDDK NIH HHS/United States
- U19 AI111825/AI/NIAID NIH HHS/United States
- R01 AI105343/AI/NIAID NIH HHS/United States
- R01 AI139119/AI/NIAID NIH HHS/United States
- T32 AR076951/AR/NIAMS NIH HHS/United States
- R01 AI123539/AI/NIAID NIH HHS/United States
- U19 AI104209/AI/NIAID NIH HHS/United States
- UM2 AI130836/AI/NIAID NIH HHS/United States
- R01 HL137006/HL/NHLBI NIH HHS/United States
- U01 DK112217/DK/NIDDK NIH HHS/United States
- U54 CA260517/CA/NCI NIH HHS/United States
- R01 HL137915/HL/NHLBI NIH HHS/United States
- T32 HL007586/HL/NHLBI NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- UM1 AI144288/AI/NIAID NIH HHS/United States
- P30 AI045008/AI/NIAID NIH HHS/United States
- UM1 AI130839/AI/NIAID NIH HHS/United States
- U01 AI150741/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

