Can functional genomic diversity provide novel insights into mechanisms of community assembly? A pilot study from an invaded alpine streambed
- PMID: 34522362
- PMCID: PMC8427620
- DOI: 10.1002/ece3.7973
Can functional genomic diversity provide novel insights into mechanisms of community assembly? A pilot study from an invaded alpine streambed
Abstract
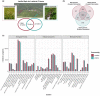
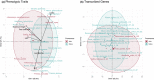
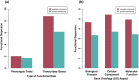
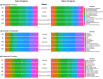
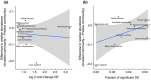
An important focus of community ecology, including invasion biology, is to investigate functional trait diversity patterns to disentangle the effects of environmental and biotic interactions. However, a notable limitation is that studies usually rely on a small and easy-to-measure set of functional traits, which might not immediately reflect ongoing ecological responses to changing abiotic or biotic conditions, including those that occur at a molecular or physiological level. We explored the potential of using the diversity of expressed genes-functional genomic diversity (FGD)-to understand ecological dynamics of a recent and ongoing alpine invasion. We quantified FGD based on transcriptomic data measured for 26 plant species occurring along adjacent invaded and pristine streambeds. We used an RNA-seq approach to summarize the overall number of expressed transcripts and their annotations to functional categories, and contrasted this with functional trait diversity (FTD) measured from a suite of characters that have been traditionally considered in plant ecology. We found greater FGD and FTD in the invaded community, independent of differences in species richness. However, the magnitude of functional dispersion was greater from the perspective of FGD than from FTD. Comparing FGD between congeneric alien-native species pairs, we did not find many significant differences in the proportion of genes whose annotations matched functional categories. Still, native species with a greater relative abundance in the invaded community compared with the pristine tended to express a greater fraction of genes at significant levels in the invaded community, suggesting that changes in FGD may relate to shifts in community composition. Comparisons of diversity patterns from the community to the species level offer complementary insights into processes and mechanisms driving invasion dynamics. FGD has the potential to illuminate cryptic changes in ecological diversity, and we foresee promising avenues for future extensions across taxonomic levels and macro-ecosystems.
Keywords: RNA‐seq; alien; alpine; community genomics; functional trait; gene expression; invasive; transcriptome.
© 2021 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to claim in the publication of this research.
Figures
References
-
- Ackerly, D. D. (2003). Community assembly, niche conservatism, and adaptive evolution in changing environments. International Journal of Plant Sciences, 164(S3), S165–S184. 10.1086/368401 - DOI
-
- Berardini, T. Z., Mundodi, S., Reiser, L., Huala, E., Garcia‐Hernandez, M., Zhang, P., Mueller, L. A., Yoon, J., Doyle, A., Lander, G., Moseyko, N., Yoo, D., Xu, I., Zoeckler, B., Montoya, M., Miller, N., Weems, D., & Rhee, S. Y. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiology, 135(2), 745–755. 10.1104/pp.104.040071 - DOI - PMC - PubMed
-
- Blanco, M. A., Whitten, W. M., Penneys, D. S., Williams, N. H., Neubig, K. M., & Endara, L. (2006). A simple and safe method for rapid drying of plant specimens using forced‐air space heaters. Selbyana, 27(1), 83–87.
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

