The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases
- PMID: 34522705
- PMCID: PMC8427257
- DOI: 10.1016/j.gendis.2020.07.011
The critical roles of m6A modification in metabolic abnormality and cardiovascular diseases
Abstract
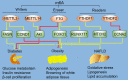
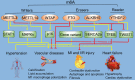
N6-methyladenosine (m6A) RNA methylation is an emerging area of epigenetics, which is a reversible and dynamic modification mediating by 'writers' (methylase, adding methyl groups, METTL3, METTL14, and WTAP), 'erasers' (demethylase, deleting methyl groups, FTO and ALKBH5), and 'readers' (YTHDF1-3, YTHDC1 and YTHDC2). Recent studies in human, animal models and cell levels have disclosed a critical role of m6A modification in regulating the homeostasis of metabolic processes and cardiovascular function. Evidence from these studies identify m6A as a candidate of biomarker and therapeutic target for metabolic abnormality and cardiovascular diseases (CVD). Comprehensive understanding of the complexity of m6A regulation in metabolic diseases and CVD will be helpful for us to understand the pathogenesis of CVD. In this review, we discuss the regulatory role of m6A in metabolic abnormality and CVD. We will emphasize the clinical relevance of m6A dysregulation in CVD.
Keywords: Cardiovascular disease; FTO; Heart failure; METTL3; Metabolic syndrome; Myocardial infarction; N6-methyladenosine; RNA epigenetics.
© 2020 Chongqing Medical University. Production and hosting by Elsevier B.V.
Figures

References
-
- Li P., Ge J., Li H. Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol. 2020;17(2):96–115. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

