Insights into the Lignocellulose-Degrading Enzyme System of Humicola grisea var. thermoidea Based on Genome and Transcriptome Analysis
- PMID: 34523973
- PMCID: PMC8557918
- DOI: 10.1128/Spectrum.01088-21
Insights into the Lignocellulose-Degrading Enzyme System of Humicola grisea var. thermoidea Based on Genome and Transcriptome Analysis
Abstract
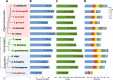
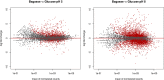
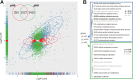
Humicola grisea var. thermoidea is a thermophilic ascomycete and important enzyme producer that has an efficient enzymatic system with a broad spectrum of thermostable carbohydrate-active (CAZy) enzymes. These enzymes can be employed in lignocellulose biomass deconstruction and other industrial applications. In this work, the genome of H. grisea var. thermoidea was sequenced. The acquired sequence reads were assembled into a total length of 28.75 Mbp. Genome features correlate with what was expected for thermophilic Sordariomycetes. The transcriptomic data showed that sugarcane bagasse significantly upregulated genes related to primary metabolism and polysaccharide deconstruction, especially hydrolases, at both pH 5 and pH 8. However, a number of exclusive and shared genes between the pH values were found, especially at pH 8. H. grisea expresses an average of 211 CAZy enzymes (CAZymes), which are capable of acting in different substrates. The top upregulated genes at both pH values represent CAZyme-encoding genes from different classes, including acetylxylan esterase, endo-1,4-β-mannosidase, exoglucanase, and endoglucanase genes. For the first time, the arsenal that the thermophilic fungus H. grisea var. thermoidea possesses to degrade the lignocellulosic biomass is shown. Carbon source and pH are of pivotal importance in regulating gene expression in this organism, and alkaline pH is a key regulatory factor for sugarcane bagasse hydrolysis. This work paves the way for the genetic manipulation and robust biotechnological applications of this fungus. IMPORTANCE Most studies regarding the use of fungi as enzyme producers for biomass deconstruction have focused on mesophile species, whereas the potential of thermophiles has been evaluated less. This study revealed, through genome and transcriptome analyses, the genetic repertoire of the biotechnological relevant thermophile fungus Humicola grisea. Comparative genomics helped us to further understand the biology and biotechnological potential of H. grisea. The results demonstrate that this fungus possesses an arsenal of carbohydrate-active (CAZy) enzymes to degrade the lignocellulosic biomass. Indeed, it expresses more than 200 genes encoding CAZy enzymes when cultivated in sugarcane bagasse. Carbon source and pH are key factors for regulating the gene expression in this organism. This work shows, for the first time, the great potential of H. grisea as an enzyme producer and a gene donor for biotechnological applications and provides the base for the genetic manipulation and robust biotechnological applications of this fungus.
Keywords: CAZy enzymes; Humicola grisea; genome sequencing; pH regulation; sugarcane bagasse; transcriptome.
Figures

References
-
- Braga M, Ferreira PM, Almeida JRM. 2020. Screening method to prioritize relevant bio-based acids and their biochemical processes using recent patent information. Biofuels, Bioprod Bioref 15:231–249.
-
- de Jong E, Stichnothe H, Bell G, Jorgensen H, de Bari I, van Haveren J, Lindorfer J. 2020. Bio-based chemicals. IEA Bioenergy.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

