Pleiotropy data resource as a primer for investigating co-morbidities/multi-morbidities and their role in disease
- PMID: 34524473
- PMCID: PMC8913486
- DOI: 10.1007/s00335-021-09917-w
Pleiotropy data resource as a primer for investigating co-morbidities/multi-morbidities and their role in disease
Abstract
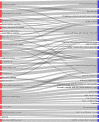
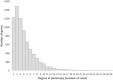
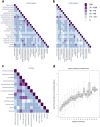
Most current biomedical and protein research focuses only on a small proportion of genes, which results in a lost opportunity to identify new gene-disease associations and explore new opportunities for therapeutic intervention. The International Mouse Phenotyping Consortium (IMPC) focuses on elucidating gene function at scale for poorly characterized and/or under-studied genes. A key component of the IMPC initiative is the implementation of a broad phenotyping pipeline, which is facilitating the discovery of pleiotropy. Characterizing pleiotropy is essential to identify gene-disease associations, and it is of particular importance when elucidating the genetic causes of syndromic disorders. Here we show how the IMPC is effectively uncovering pleiotropy and how the new mouse models and gene function hypotheses generated by the IMPC are increasing our understanding of the mammalian genome, forming the basis of new research and identifying new gene-disease associations.
© 2021. The Author(s).
Conflict of interest statement
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Figures

References
-
- Albert AY, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62:76–85. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

