Historical, current, and emerging tools for identification and serotyping of Shigella
- PMID: 34524650
- PMCID: PMC8441030
- DOI: 10.1007/s42770-021-00573-5
Historical, current, and emerging tools for identification and serotyping of Shigella
Abstract
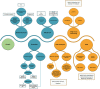
The Shigella genus includes serious foodborne disease etiologic agents, with 4 species and 54 serotypes. Identification at species and serotype levels is a crucial task in microbiological laboratories. Nevertheless, the genetic similarity between Shigella spp. and Escherichia coli challenges the correct identification and serotyping of Shigella spp., with subsequent negative repercussions on surveillance, epidemiological investigations, and selection of appropriate treatments. For this purpose, multiple techniques have been developed historically ranging from phenotype-based methods and single or multilocus molecular techniques to whole-genome sequencing (WGS). To facilitate the selection of the most relevant method, we herein provide a global overview of historical and emerging identification and serotyping techniques with a particular focus on the WGS-based approaches. This review highlights the excellent discriminatory power of WGS to more accurately elucidate the epidemiology of Shigella spp., disclose novel promising genomic targets for surveillance methods, and validate previous well-established methods.
Keywords: Escherichia coli; Identification; Serotyping; Shigella spp.; Whole-genome sequencing.
© 2021. Sociedade Brasileira de Microbiologia.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

