The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation
- PMID: 34525179
- PMCID: PMC8796651
- DOI: 10.1182/blood.2021013201
The STAT3-MYC axis promotes survival of leukemia stem cells by regulating SLC1A5 and oxidative phosphorylation
Abstract
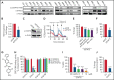
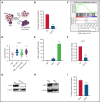
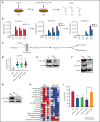
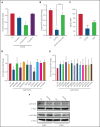
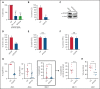
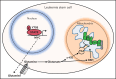
Acute myeloid leukemia (AML) is characterized by the presence of leukemia stem cells (LSCs), and failure to fully eradicate this population contributes to disease persistence/relapse. Prior studies have characterized metabolic vulnerabilities of LSCs, which demonstrate preferential reliance on oxidative phosphorylation (OXPHOS) for energy metabolism and survival. In the present study, using both genetic and pharmacologic strategies in primary human AML specimens, we show that signal transducer and activator of transcription 3 (STAT3) mediates OXPHOS in LSCs. STAT3 regulates AML-specific expression of MYC, which in turn controls transcription of the neutral amino acid transporter gene SLC1A5. We show that genetic inhibition of MYC or SLC1A5 acts to phenocopy the impairment of OXPHOS observed with STAT3 inhibition, thereby establishing this axis as a regulatory mechanism linking STAT3 to energy metabolism. Inhibition of SLC1A5 reduces intracellular levels of glutamine, glutathione, and multiple tricarboxylic acid (TCA) cycle metabolites, leading to reduced TCA cycle activity and inhibition of OXPHOS. Based on these findings, we used a novel small molecule STAT3 inhibitor, which binds STAT3 and disrupts STAT3-DNA, to evaluate the biological role of STAT3. We show that STAT3 inhibition selectively leads to cell death in AML stem and progenitor cells derived from newly diagnosed patients and patients who have experienced relapse while sparing normal hematopoietic cells. Together, these findings establish a STAT3-mediated mechanism that controls energy metabolism and survival in primitive AML cells.
© 2022 by The American Society of Hematology.
Figures
References
-
- Shah A, Andersson TM, Rachet B, Björkholm M, Lambert PC. Survival and cure of acute myeloid leukaemia in England, 1971-2006: a population-based study. Br J Haematol. 2013;162(4):509-516. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7-34. - PubMed
-
- Eppert K, Takenaka K, Lechman ER, et al. . Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086-1093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

