Increased fidelity of protein synthesis extends lifespan
- PMID: 34525330
- PMCID: PMC8570412
- DOI: 10.1016/j.cmet.2021.08.017
Increased fidelity of protein synthesis extends lifespan
Abstract
Loss of proteostasis is a fundamental process driving aging. Proteostasis is affected by the accuracy of translation, yet the physiological consequence of having fewer protein synthesis errors during multi-cellular organismal aging is poorly understood. Our phylogenetic analysis of RPS23, a key protein in the ribosomal decoding center, uncovered a lysine residue almost universally conserved across all domains of life, which is replaced by an arginine in a small number of hyperthermophilic archaea. When introduced into eukaryotic RPS23 homologs, this mutation leads to accurate translation, as well as heat shock resistance and longer life, in yeast, worms, and flies. Furthermore, we show that anti-aging drugs such as rapamycin, Torin1, and trametinib reduce translation errors, and that rapamycin extends further organismal longevity in RPS23 hyperaccuracy mutants. This implies a unified mode of action for diverse pharmacological anti-aging therapies. These findings pave the way for identifying novel translation accuracy interventions to improve aging.
Keywords: RPS23; aging; archaea; mTOR; protein synthesis; proteostasis; ribosome; translation; translation accuracy; translation fidelity.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

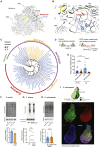
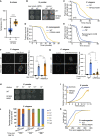
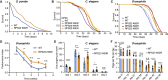

Comment in
-
Longevity lost in translation.Nat Rev Mol Cell Biol. 2021 Nov;22(11):711. doi: 10.1038/s41580-021-00422-1. Nat Rev Mol Cell Biol. 2021. PMID: 34545217 No abstract available.
-
Haste makes waste: The significance of translation fidelity for development and longevity.Mol Cell. 2021 Sep 16;81(18):3675-3676. doi: 10.1016/j.molcel.2021.08.036. Mol Cell. 2021. PMID: 34547232 Free PMC article.
-
Thermophiles reveal the clues to longevity: precise protein synthesis.J Cardiovasc Aging. 2022 Apr;2(2):14. doi: 10.20517/jca.2021.38. Epub 2022 Jan 18. J Cardiovasc Aging. 2022. PMID: 36778790 Free PMC article.
References
-
- Agarwal D., Gregory S.T., O’Connor M. Error-prone and error-restrictive mutations affecting ribosomal protein S12. J. Mol. Biol. 2011;410:1–9. - PubMed
-
- Arnold A., Rahman M.M., Lee M.C., Muehlhaeusser S., Katic I., Gaidatzis D., Hess D., Scheckel C., Wright J.E., Stetak A. Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014;42:13353–13369. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- MC_UP_1605/6/MRC_/Medical Research Council/United Kingdom
- R01 GM135599/GM/NIGMS NIH HHS/United States
- C7893/A28990/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_1102/6/MRC_/Medical Research Council/United Kingdom
- 201487 /WT_/Wellcome Trust/United Kingdom
- 102532/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- BB/V006916/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 102531/Z/13/A/WT_/Wellcome Trust/United Kingdom
- MC-A654-5QC80/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- C416/A25145/CRUK_/Cancer Research UK/United Kingdom
- 28990/CRUK_/Cancer Research UK/United Kingdom
- MR/M02492X/1/MRC_/Medical Research Council/United Kingdom
- R01 GM141694/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

