Adenosine-to-inosine editing of endogenous Z-form RNA by the deaminase ADAR1 prevents spontaneous MAVS-dependent type I interferon responses
- PMID: 34525337
- PMCID: PMC8459395
- DOI: 10.1016/j.immuni.2021.08.011
Adenosine-to-inosine editing of endogenous Z-form RNA by the deaminase ADAR1 prevents spontaneous MAVS-dependent type I interferon responses
Abstract
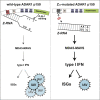
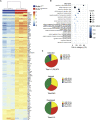
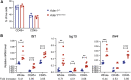
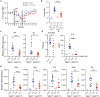
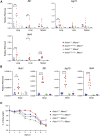
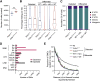
Nucleic acids are powerful triggers of innate immunity and can adopt the Z-conformation, an unusual left-handed double helix. Here, we studied the biological function(s) of Z-RNA recognition by the adenosine deaminase ADAR1, mutations in which cause Aicardi-Goutières syndrome. Adar1mZα/mZα mice, bearing two point mutations in the Z-nucleic acid binding (Zα) domain that abolish Z-RNA binding, displayed spontaneous induction of type I interferons (IFNs) in multiple organs, including in the lung, where both stromal and hematopoietic cells showed IFN-stimulated gene (ISG) induction. Lung neutrophils expressed ISGs induced by the transcription factor IRF3, indicating an initiating role for neutrophils in this IFN response. The IFN response in Adar1mZα/mZα mice required the adaptor MAVS, implicating cytosolic RNA sensing. Adenosine-to-inosine changes were enriched in transposable elements and revealed a specific requirement of ADAR1's Zα domain in editing of a subset of RNAs. Thus, endogenous RNAs in Z-conformation have immunostimulatory potential curtailed by ADAR1, with relevance to autoinflammatory disease in humans.
Keywords: ADAR1; Aicardi–Goutières syndrome; MAVS; MDA5; RNA editing; Z-RNA; Zα domain; influenza A virus; interferon; neutrophil.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


Comment in
-
ADAR1 edits the SenZ and SenZ-ability of RNA.Immunity. 2021 Sep 14;54(9):1909-1911. doi: 10.1016/j.immuni.2021.08.021. Immunity. 2021. PMID: 34525334
References
-
- Athanasiadis A. Zalpha-domains: at the intersection between RNA editing and innate immunity. Semin. Cell Dev. Biol. 2012;23:275–280. - PubMed
-
- Athanasiadis A., Placido D., Maas S., Brown B.A., 2nd, Lowenhaupt K., Rich A. The crystal structure of the Zbeta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 2005;351:496–507. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

