Loss of Christianson Syndrome Na+/H+ Exchanger 6 (NHE6) Causes Abnormal Endosome Maturation and Trafficking Underlying Lysosome Dysfunction in Neurons
- PMID: 34526390
- PMCID: PMC8570832
- DOI: 10.1523/JNEUROSCI.1244-20.2021
Loss of Christianson Syndrome Na+/H+ Exchanger 6 (NHE6) Causes Abnormal Endosome Maturation and Trafficking Underlying Lysosome Dysfunction in Neurons
Abstract
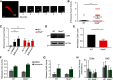
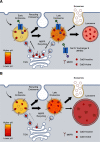
Loss-of-function mutations in endosomal Na+/H+ exchanger 6 (NHE6) cause the X-linked neurologic disorder Christianson syndrome. Patients exhibit symptoms associated with both neurodevelopmental and neurodegenerative abnormalities. While loss of NHE6 has been shown to overacidify the endosome lumen, and is associated with endolysosome neuropathology, NHE6-mediated mechanisms in endosome trafficking and lysosome function have been understudied. Here, we show that NHE6-null mouse neurons demonstrate worsening lysosome function with time in culture, likely as a result of defective endosome trafficking. NHE6-null neurons exhibit overall reduced lysosomal proteolysis despite overacidification of the endosome and lysosome lumen. Akin to Nhx1 mutants in Saccharomyces cerevisiae, we observe decreased endosome-lysosome fusion in NHE6-null neurons. Also, we find premature activation of pH-dependent cathepsin D (CatD) in endosomes. While active CatD is increased in endosomes, CatD activation and CatD protein levels are reduced in the lysosome. Protein levels of another mannose 6-phosphate receptor (M6PR)-dependent enzyme, β-N-acetylglucosaminidase, were also decreased in lysosomes of NHE6-null neurons. M6PRs accumulate in late endosomes, suggesting defective M6PR recycling and retromer function in NHE6-null neurons. Finally, coincident with decreased endosome-lysosome fusion, using total internal reflection fluorescence, we also find a prominent increase in fusion between endosomal multivesicular bodies and the plasma membrane, indicating enhanced exosome secretion from NHE6-null neurons. In summary, in addition to overacidification of endosomes and lysosomes, loss of NHE6 leads to defects in endosome maturation and trafficking, including enhanced exosome release, contributing to lysosome deficiency and potentially leading to neurodegenerative disease.SIGNIFICANCE STATEMENT Loss-of-function mutations in the endosomal Na+/H+ exchanger 6 (NHE6) cause Christianson syndrome, an X-linked neurologic disorder. Loss of NHE6 has been shown to overacidify endosomes; however, endosome trafficking mechanisms have been understudied, and the mechanisms leading to neurodegeneration are largely unknown. In NHE6-null mouse neurons in vitro, we find worsening lysosome function with days in culture. Notably, pH-dependent lysosome enzymes, such as cathepsin D, have reduced activity in lysosomes yet increased, precocious activity in endosomes in NHE6-null neurons. Further, endosomes show reduced fusion to lysosomes, and increased fusion to the plasma membrane with increased exosome release. This study identifies new mechanisms involving defective endosome maturation and trafficking that impair lysosome function in Christianson syndrome, likely contributing to neurodegeneration.
Keywords: Christianson syndrome; NHE6; endosome; exosome; lysosome; neurodegeneration.
Copyright © 2021 Pescosolido et al.
Figures







References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
