Rapid MRSA detection via tandem mass spectrometry of the intact 80 kDa PBP2a resistance protein
- PMID: 34526615
- PMCID: PMC8443585
- DOI: 10.1038/s41598-021-97844-w
Rapid MRSA detection via tandem mass spectrometry of the intact 80 kDa PBP2a resistance protein
Abstract
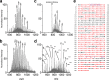
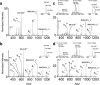
Treatment of antibiotic-resistant infections is dependent on the detection of specific bacterial genes or proteins in clinical assays. Identification of methicillin-resistant Staphylococcus aureus (MRSA) is often accomplished through the detection of penicillin-binding protein 2a (PBP2a). With greater dependence on mass spectrometry (MS)-based bacterial identification, complementary efforts to detect resistance have been hindered by the complexity of those proteins responsible. Initial characterization of PBP2a indicates the presence of glycan modifications. To simplify detection, we demonstrate a proof-of-concept tandem MS approach involving the generation of N-terminal PBP2a peptide-like fragments and detection of unique product ions during top-down proteomic sample analyses. This approach was implemented for two PBP2a variants, PBP2amecA and PBP2amecC, and was accurate across a representative panel of MRSA strains with different genetic backgrounds. Additionally, PBP2amecA was successfully detected from clinical isolates using a five-minute liquid chromatographic separation and implementation of this MS detection strategy. Our results highlight the capability of direct MS-based resistance marker detection and potential advantages for implementing these approaches in clinical diagnostics.
© 2021. The Author(s).
Conflict of interest statement
J.R.N., A.V., S.R.K., W.M.M., C.M., M.V., A.K., H.F., J.F., J.R., J.E.P.S., and J.L.S. are employees of Thermo Fisher Scientific. J.R.N., W.M.M., J.E.P.S. and J.L.S. all own company stock. J.R.N., W.M.M., J.F., and J.L.S. are listed as authors on patents describing the SPE tip or method descriptions in this manuscript.
Figures


References
-
- CDC. Antibiotic Resistance Threats in the United States, 2019. 150 (U.S. Department of Health and Human Services, 2019).
-
- Lim D, Strynadka NCJ. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 2002;9:870–876. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

