Single-molecule kinetic locking allows fluorescence-free quantification of protein/nucleic-acid binding
- PMID: 34526657
- PMCID: PMC8443601
- DOI: 10.1038/s42003-021-02606-z
Single-molecule kinetic locking allows fluorescence-free quantification of protein/nucleic-acid binding
Abstract
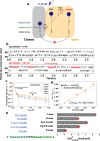
Fluorescence-free micro-manipulation of nucleic acids (NA) allows the functional characterization of DNA/RNA processing proteins, without the interference of labels, but currently fails to detect and quantify their binding. To overcome this limitation, we developed a method based on single-molecule force spectroscopy, called kinetic locking, that allows a direct in vitro visualization of protein binding while avoiding any kind of chemical disturbance of the protein's natural function. We validate kinetic locking by measuring accurately the hybridization energy of ultrashort nucleotides (5, 6, 7 bases) and use it to measure the dynamical interactions of Escherichia coli/E. coli RecQ helicase with its DNA substrate.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Sudhaker, S. et al. Germanium nanospheres for ultraresolution picotensiometry of kinesin motors. Science10.1126/science.abd9944 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ANR-14-CE10-0014/Agence Nationale de la Recherche (French National Research Agency)
- ANR-19-CE11-0021-01/Agence Nationale de la Recherche (French National Research Agency)
- [267 862]/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

