Multi-omics mapping of human papillomavirus integration sites illuminates novel cervical cancer target genes
- PMID: 34526665
- PMCID: PMC8575955
- DOI: 10.1038/s41416-021-01545-0
Multi-omics mapping of human papillomavirus integration sites illuminates novel cervical cancer target genes
Abstract
Background: Integration of human papillomavirus (HPV) into the host genome is a dominant feature of invasive cervical cancer (ICC), yet the tumorigenicity of cis genomic changes at integration sites remains largely understudied.
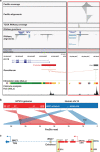
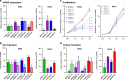
Methods: Combining multi-omics data from The Cancer Genome Atlas with patient-matched long-read sequencing of HPV integration sites, we developed a strategy for using HPV integration events to identify and prioritise novel candidate ICC target genes (integration-detected genes (IDGs)). Four IDGs were then chosen for in vitro functional studies employing small interfering RNA-mediated knockdown in cell migration, proliferation and colony formation assays.
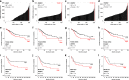
Results: PacBio data revealed 267 unique human-HPV breakpoints comprising 87 total integration events in eight tumours. Candidate IDGs were filtered based on the following criteria: (1) proximity to integration site, (2) clonal representation of integration event, (3) tumour-specific expression (Z-score) and (4) association with ICC survival. Four candidates prioritised based on their unknown function in ICC (BNC1, RSBN1, USP36 and TAOK3) exhibited oncogenic properties in cervical cancer cell lines. Further, annotation of integration events provided clues regarding potential mechanisms underlying altered IDG expression in both integrated and non-integrated ICC tumours.
Conclusions: HPV integration events can guide the identification of novel IDGs for further study in cervical carcinogenesis and as putative therapeutic targets.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures