Respiratory Microbiota Profiles Associated With the Progression From Airway Inflammation to Remodeling in Mice With OVA-Induced Asthma
- PMID: 34526979
- PMCID: PMC8435892
- DOI: 10.3389/fmicb.2021.723152
Respiratory Microbiota Profiles Associated With the Progression From Airway Inflammation to Remodeling in Mice With OVA-Induced Asthma
Abstract
Background: The dysbiosis of respiratory microbiota plays an important role in asthma development. However, there is limited information on the changes in the respiratory microbiota and how these affect the host during the progression from acute allergic inflammation to airway remodeling in asthma.
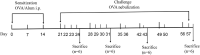
Objective: An ovalbumin (OVA)-induced mouse model of chronic asthma was established to explore the dynamic changes in the respiratory microbiota in the different stages of asthma and their association with chronic asthma progression.
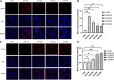
Methods: Hematoxylin and eosin (H&E), periodic acid-schiff (PAS), and Masson staining were performed to observe the pathological changes in the lung tissues of asthmatic mice. The respiratory microbiota was analyzed using 16S rRNA gene sequencing followed by taxonomical analysis. The cytokine levels in bronchoalveolar lavage fluid (BALF) specimens were measured. The matrix metallopeptidase 9 (MMP-9) and vascular endothelial growth factor (VEGF-A) expression levels in lung tissues were measured to detect airway remodeling in OVA-challenged mice.
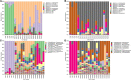
Results: Acute allergic inflammation was the major manifestation at weeks 1 and 2 after OVA atomization stimulation, whereas at week 6 after the stimulation, airway remodeling was the most prominent observation. In the acute inflammatory stage, Pseudomonas was more abundant, whereas Staphylococcus and Cupriavidus were more abundant at the airway remodeling stage. The microbial compositions of the upper and lower respiratory tracts were similar. However, the dominant respiratory microbiota in the acute inflammatory and airway remodeling phases were different. Metagenomic functional prediction showed that the pathways significantly upregulated in the acute inflammatory phase and airway remodeling phase were different. The cytokine levels in BALF and the expression patterns of proteins associated with airway remodeling in the lung tissue were consistent with the metagenomic function results.
Conclusion: The dynamic changes in respiratory microbiota are closely associated with the progression of chronic asthma. Metagenomic functional prediction indicated the changes associated with acute allergic inflammation and airway remodeling.
Keywords: airway inflammation; airway remodeling; asthma; metagenomic functional prediction; respiratory microbiota.
Copyright © 2021 Zheng, Wu, Zou, Wang, He and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

