Identification of Tumor Antigens and Immune Landscape in Glioblastoma for mRNA Vaccine Development
- PMID: 34527020
- PMCID: PMC8435740
- DOI: 10.3389/fgene.2021.701065
Identification of Tumor Antigens and Immune Landscape in Glioblastoma for mRNA Vaccine Development
Abstract
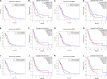
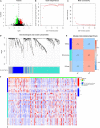
Background: Clinical benefits from standard therapies against glioblastoma (GBM) are limited in part due to the intrinsic radio- and chemo-resistance. As an essential part of tumor immunotherapy for adjunct, therapeutic tumor vaccines have been effective against multiple solid cancers, while their efficacy against GBM remains undefined. Therefore, this study aims to find the possible tumor antigens of GBM and identify the suitable population for cancer vaccination through immunophenotyping. Method: The genomic and responding clinical data of 169 GBM samples and five normal brain samples were obtained from The Cancer Genome Atlas (TCGA). The mRNA_seq data of 940 normal brain tissue were downloaded from Genotype-Tissue Expression (GTEx). Potential GBM mRNA antigens were screened out by differential expression, copy number variant (CNV), and mutation analysis. K-M survival and Cox analysis were carried out to investigate the prognostic association of potential tumor antigens. Tumor Immune Estimation Resource (TIMER) was used to explore the association between the antigens and tumor immune infiltrating cells (TIICs). Immunophenotyping of 169 samples was performed through consensus clustering based on the abundance of 22 kinds of immune cells. The characteristics of the tumor immune microenvironment (TIME) in each cluster were explored through single-sample gene set enrichment analysis based on 29 kinds of immune-related hallmarks and pathways. Weighted gene co-expression network analysis (WGCNA) was performed to cluster the genes related to immune subtypes. Finally, pathway enrichment analyses were performed to annotate the potential function of modules screened through WGCNA. Results: Two potential tumor antigens selected were significantly positively associated with the antigen-presenting immune cells (APCs) in GBM. Furthermore, the expression of antigens was verified at the protein level by Immunohistochemistry. Two robust immune subtypes, immune subtype 1 (IS1) and immune subtype 2 (IS2), representing immune status "immune inhibition" and "immune inflamed", respectively, had distinct clinical outcomes in GBM. Conclusion: ARPC1B and HK3 were potential mRNA antigens for developing GBM mRNA vaccination, and the patients in IS2 were considered the most suitable population for vaccination in GBM.
Keywords: bioinformatics; cancer vaccination; glioblastoma; immunophenotyping; tumor antigens.
Copyright © 2021 Ye, Wang, Yang, Hu, Zhang, Tong, Liu and Tian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

